Tree building and visualization
Source:vignettes/articles/tree_visualization.Rmd
tree_visualization.RmdIn introduction, you can read the review of (zou2024?), entitled “Common Methods for Phylogenetic Tree Construction and Their Implementation in R“.
library("tidytree") # first load to disable warning about phylo class
library("MiscMetabar")
library("phangorn")
data(data_fungi)
df <- subset_taxa_pq(data_fungi, taxa_sums(data_fungi) > 9000)
df_tree <- quiet(build_phytree_pq(df, nb_bootstrap = 5))
data_fungi_tree <- merge_phyloseq(df, phyloseq::phy_tree(df_tree$ML$tree))
library("treeio")
library("ggtree")
ggtree(data_fungi_tree@phy_tree, layout = "ellipse") + geom_tiplab()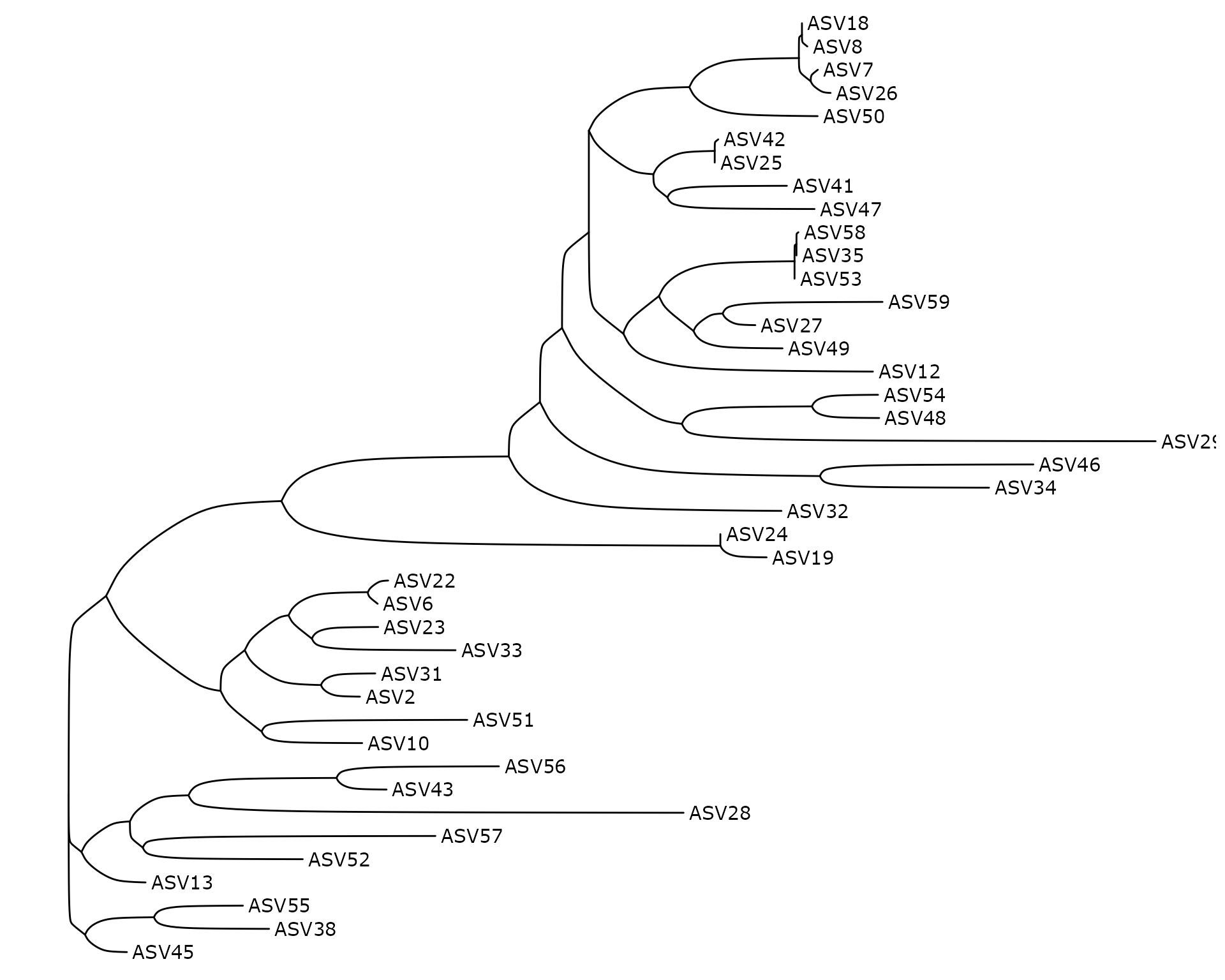
ggtree(as.treedata(df_tree$ML), layout = "slanted")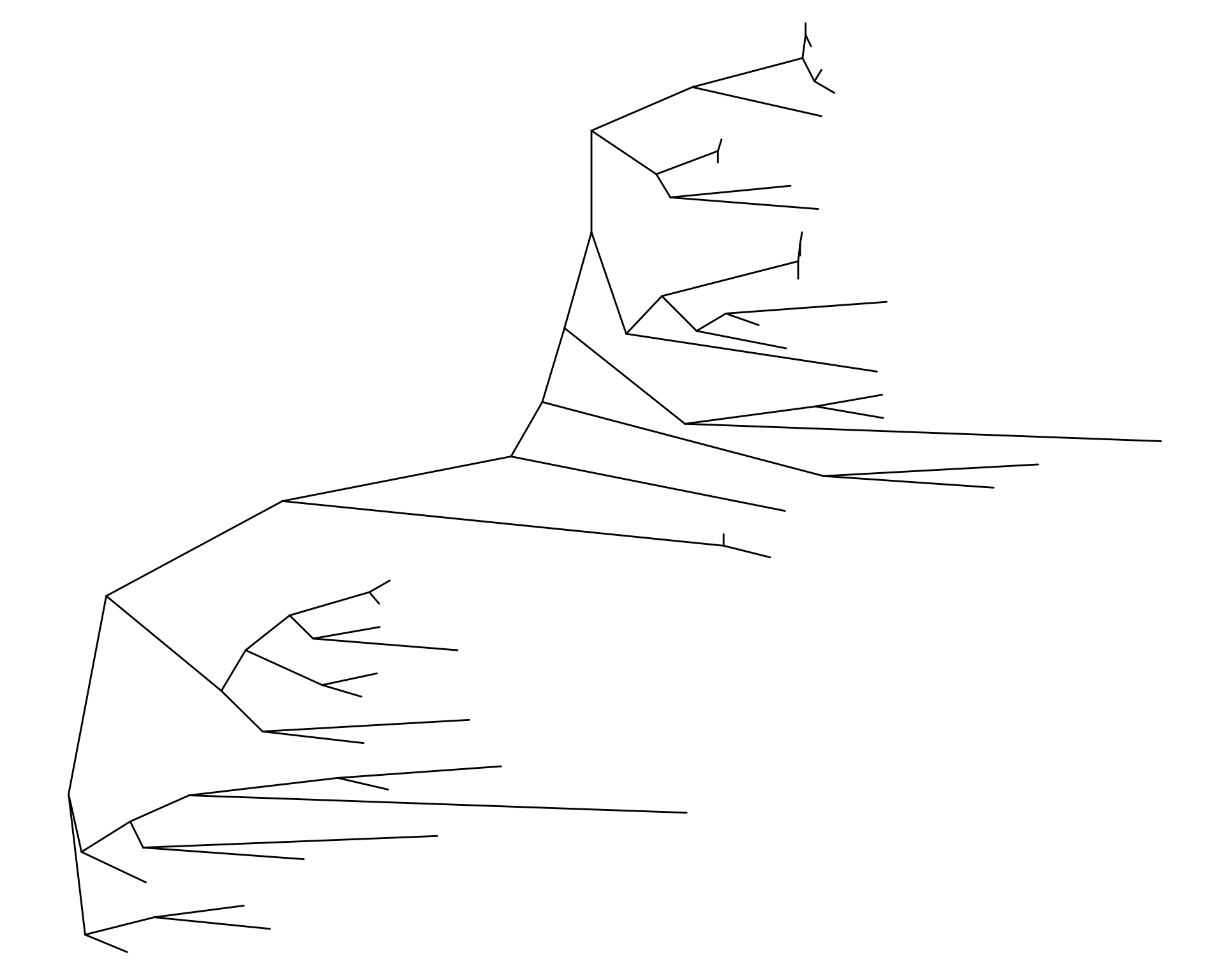
ggdensitree(df_tree$ML_bs, alpha = .3, colour = "steelblue") +
geom_tiplab(size = 3) + hexpand(.35)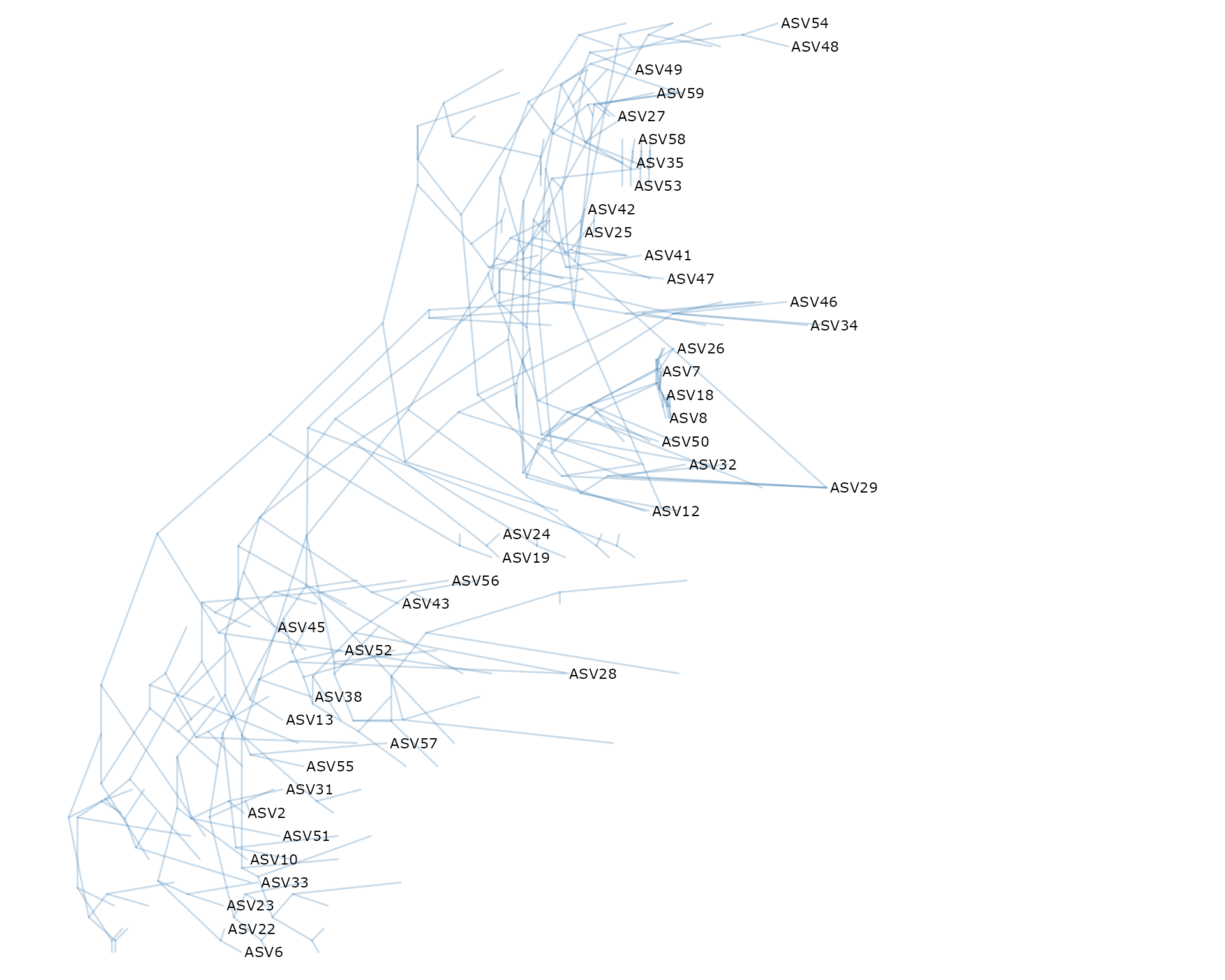
ggtree(as.treedata(df_tree$ML)) +
geom_text(aes(x = branch, label = AA_subs, vjust = -.5), size = 1)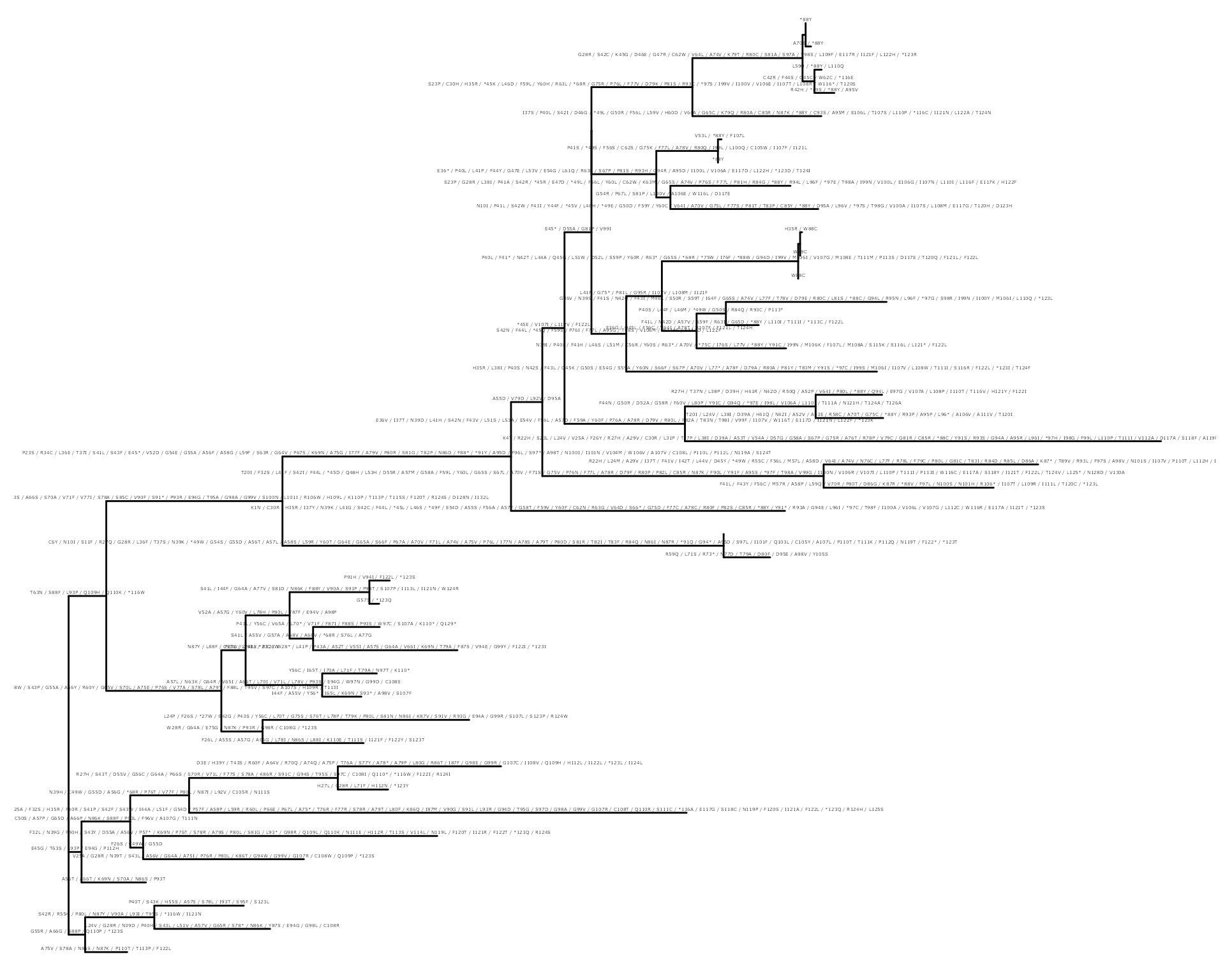
tax_tab <- as.data.frame(data_fungi_tree@tax_table)
tax_tab <- data.frame("OTU" = rownames(tax_tab), tax_tab)
p <- ggtree(as.treedata(data_fungi_tree@phy_tree)) %<+%
tax_tab
p + geom_tippoint(aes(color = Class, shape = Phylum)) +
geom_text(aes(label = Genus), hjust = -0.2, size = 2)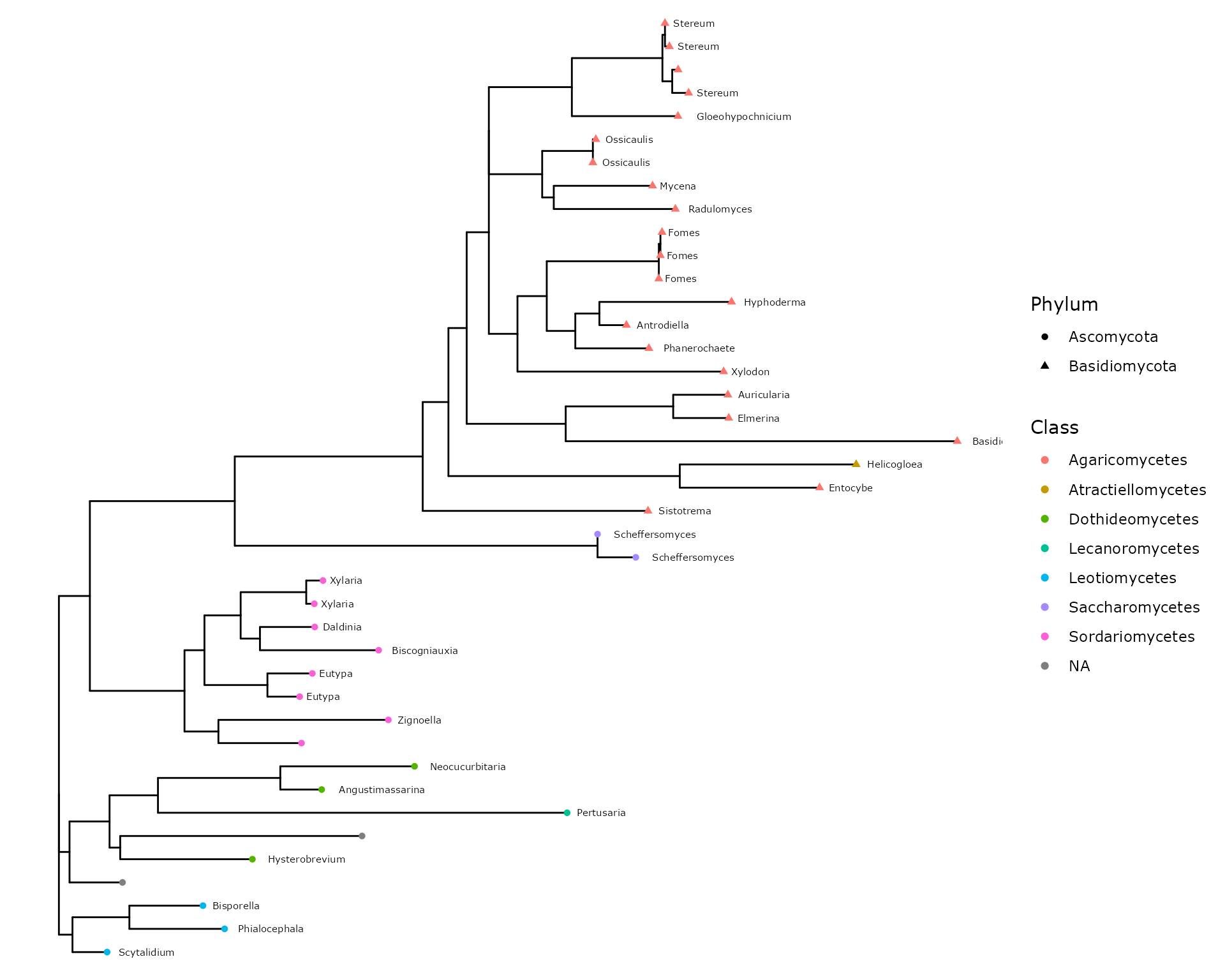
ggtree(as.treedata(data_fungi_tree@phy_tree), branch.length = "none") %<+%
tax_tab +
geom_tippoint(aes(color = Class, shape = Phylum), size = 2)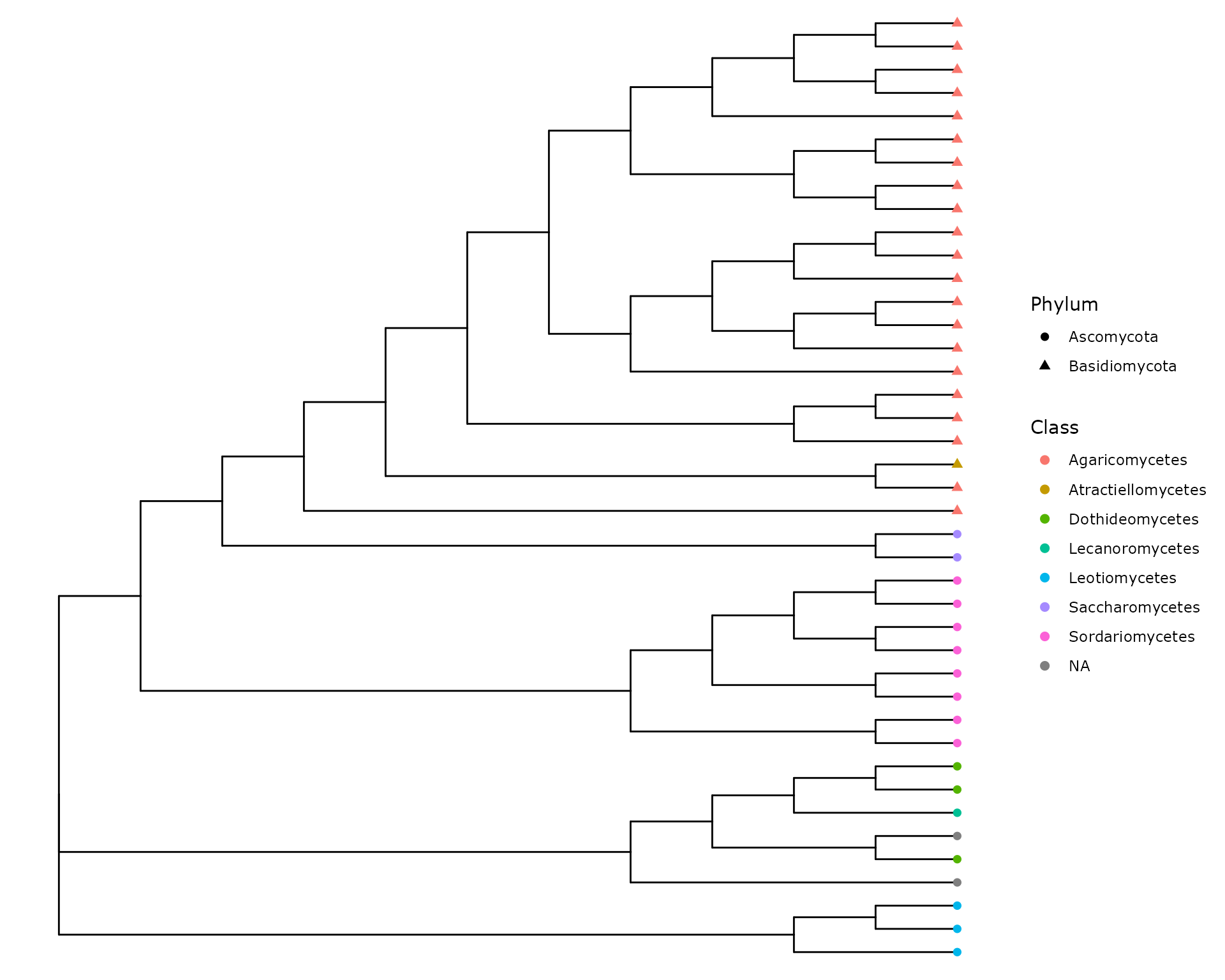
Session information
sessionInfo()
#> R version 4.5.1 (2025-06-13)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Kali GNU/Linux Rolling
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.29.so; LAPACK version 3.12.0
#>
#> locale:
#> [1] LC_CTYPE=fr_FR.UTF-8 LC_NUMERIC=C
#> [3] LC_TIME=fr_FR.UTF-8 LC_COLLATE=fr_FR.UTF-8
#> [5] LC_MONETARY=fr_FR.UTF-8 LC_MESSAGES=fr_FR.UTF-8
#> [7] LC_PAPER=fr_FR.UTF-8 LC_NAME=C
#> [9] LC_ADDRESS=C LC_TELEPHONE=C
#> [11] LC_MEASUREMENT=fr_FR.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: Europe/Paris
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ggtree_3.16.3 treeio_1.32.0 phangorn_2.12.1 ape_5.8-1
#> [5] MiscMetabar_0.14.4 purrr_1.1.0 dplyr_1.1.4 dada2_1.36.0
#> [9] Rcpp_1.1.0 ggplot2_4.0.0 phyloseq_1.52.0 tidytree_0.4.6
#>
#> loaded via a namespace (and not attached):
#> [1] DBI_1.2.3 bitops_1.0-9
#> [3] deldir_2.0-4 permute_0.9-8
#> [5] rlang_1.1.6 magrittr_2.0.4
#> [7] ade4_1.7-23 matrixStats_1.5.0
#> [9] compiler_4.5.1 mgcv_1.9-3
#> [11] png_0.1-8 systemfonts_1.2.3
#> [13] vctrs_0.6.5 reshape2_1.4.4
#> [15] quadprog_1.5-8 stringr_1.5.2
#> [17] pwalign_1.4.0 pkgconfig_2.0.3
#> [19] crayon_1.5.3 fastmap_1.2.0
#> [21] XVector_0.48.0 labeling_0.4.3
#> [23] Rsamtools_2.24.1 rmarkdown_2.29
#> [25] UCSC.utils_1.4.0 ragg_1.5.0
#> [27] xfun_0.53 cachem_1.1.0
#> [29] aplot_0.2.9 GenomeInfoDb_1.44.3
#> [31] jsonlite_2.0.0 biomformat_1.36.0
#> [33] rhdf5filters_1.20.0 DelayedArray_0.34.1
#> [35] Rhdf5lib_1.30.0 BiocParallel_1.42.2
#> [37] jpeg_0.1-11 parallel_4.5.1
#> [39] cluster_2.1.8.1 R6_2.6.1
#> [41] bslib_0.9.0 stringi_1.8.7
#> [43] RColorBrewer_1.1-3 GenomicRanges_1.60.0
#> [45] jquerylib_0.1.4 SummarizedExperiment_1.38.1
#> [47] iterators_1.0.14 knitr_1.50
#> [49] DECIPHER_3.4.0 IRanges_2.42.0
#> [51] Matrix_1.7-4 splines_4.5.1
#> [53] igraph_2.1.4 tidyselect_1.2.1
#> [55] abind_1.4-8 yaml_2.3.10
#> [57] vegan_2.7-1 codetools_0.2-20
#> [59] hwriter_1.3.2.1 lattice_0.22-7
#> [61] tibble_3.3.0 plyr_1.8.9
#> [63] Biobase_2.68.0 withr_3.0.2
#> [65] ShortRead_1.66.0 S7_0.2.0
#> [67] evaluate_1.0.5 gridGraphics_0.5-1
#> [69] desc_1.4.3 survival_3.8-3
#> [71] RcppParallel_5.1.11-1 Biostrings_2.76.0
#> [73] pillar_1.11.1 MatrixGenerics_1.20.0
#> [75] foreach_1.5.2 stats4_4.5.1
#> [77] ggfun_0.2.0 generics_0.1.4
#> [79] S4Vectors_0.46.0 scales_1.4.0
#> [81] glue_1.8.0 lazyeval_0.2.2
#> [83] tools_4.5.1 interp_1.1-6
#> [85] data.table_1.17.8 GenomicAlignments_1.44.0
#> [87] fs_1.6.6 fastmatch_1.1-6
#> [89] rhdf5_2.52.1 grid_4.5.1
#> [91] tidyr_1.3.1 latticeExtra_0.6-31
#> [93] patchwork_1.3.2 nlme_3.1-168
#> [95] GenomeInfoDbData_1.2.14 cli_3.6.5
#> [97] rappdirs_0.3.3 textshaping_1.0.3
#> [99] S4Arrays_1.8.1 gtable_0.3.6
#> [101] yulab.utils_0.2.1 sass_0.4.10
#> [103] digest_0.6.37 BiocGenerics_0.54.0
#> [105] ggplotify_0.1.3 SparseArray_1.8.1
#> [107] htmlwidgets_1.6.4 farver_2.1.2
#> [109] htmltools_0.5.8.1 pkgdown_2.1.3
#> [111] multtest_2.64.0 lifecycle_1.0.4
#> [113] httr_1.4.7 MASS_7.3-65