summarytools::ctable(sam_tab$Height, sam_tab$Time, chisq = TRUE)
#> Warning in chisq.test(freq_table_min): Chi-squared approximation may be
#> incorrect
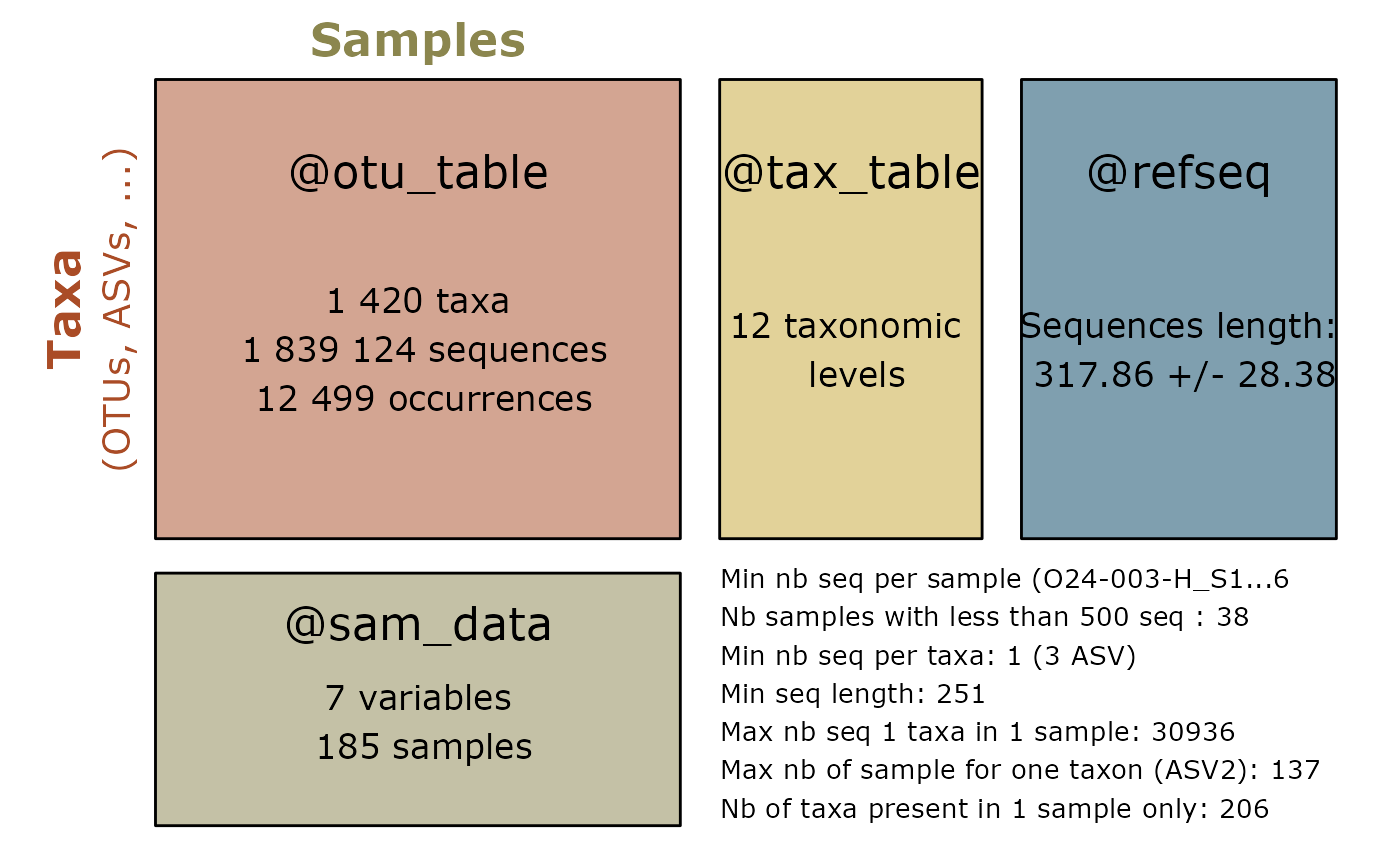
pillar::glimpse(sam_tab)
#> Warning in class(x) <- tibble_class: Setting class(x) to multiple strings
#> ("tbl_df", "tbl", ...); result will no longer be an S4 object
report::report_sample(data_fungi@sam_data, select = c("Height", "Time"), by = "Tree_name")
pointblank::scan_data(sam_tab, sections = "OVSCMI")
#> Warning in class(x) <- tibble_class: Setting class(x) to multiple strings
#> ("tbl_df", "tbl", ...); result will no longer be an S4 object
pointblank::create_informant(
tbl = sam_tab,
tbl_name = "Sample data",
label = "data_fungi"
)
Explore taxonomic data
summarytools::ctable(grepl("Ectomycorrhizal", tax_tab$Guild), tax_tab$Class, chisq = TRUE)
#> Warning in chisq.test(freq_table_min): Chi-squared approximation may be
#> incorrect
datawizard::data_codebook(tax_tab)
#> Warning: The table contains very wide columns that don't fit into the available
#> display-width of the console. Splitting tables into multiple parts did
#> not have the desired effect.
pointblank::scan_data(tax_tab, sections = "OVSCM")
pointblank::create_informant(
tbl = tax_tab,
tbl_name = "Taxonomic table",
label = "data_fungi"
)
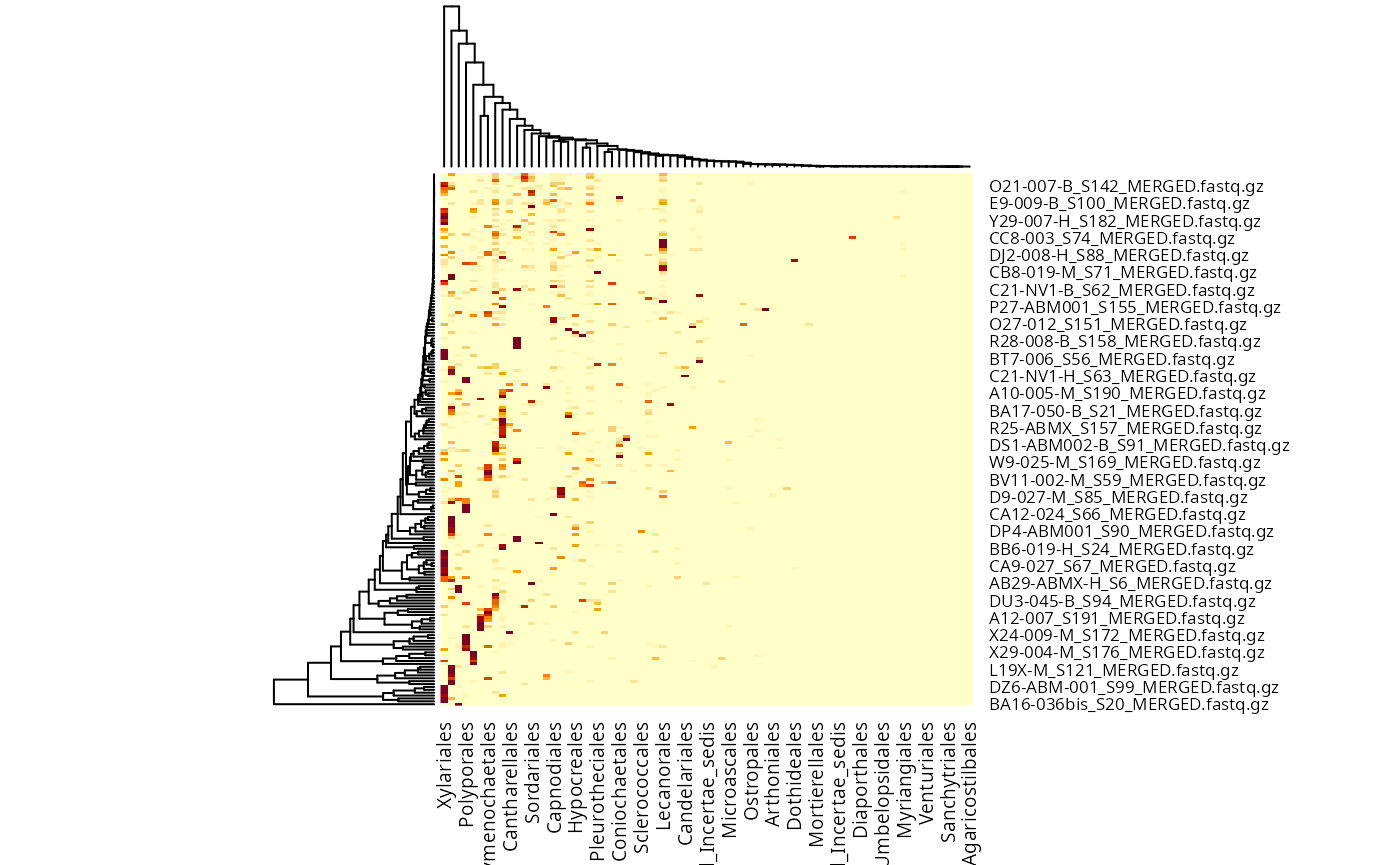
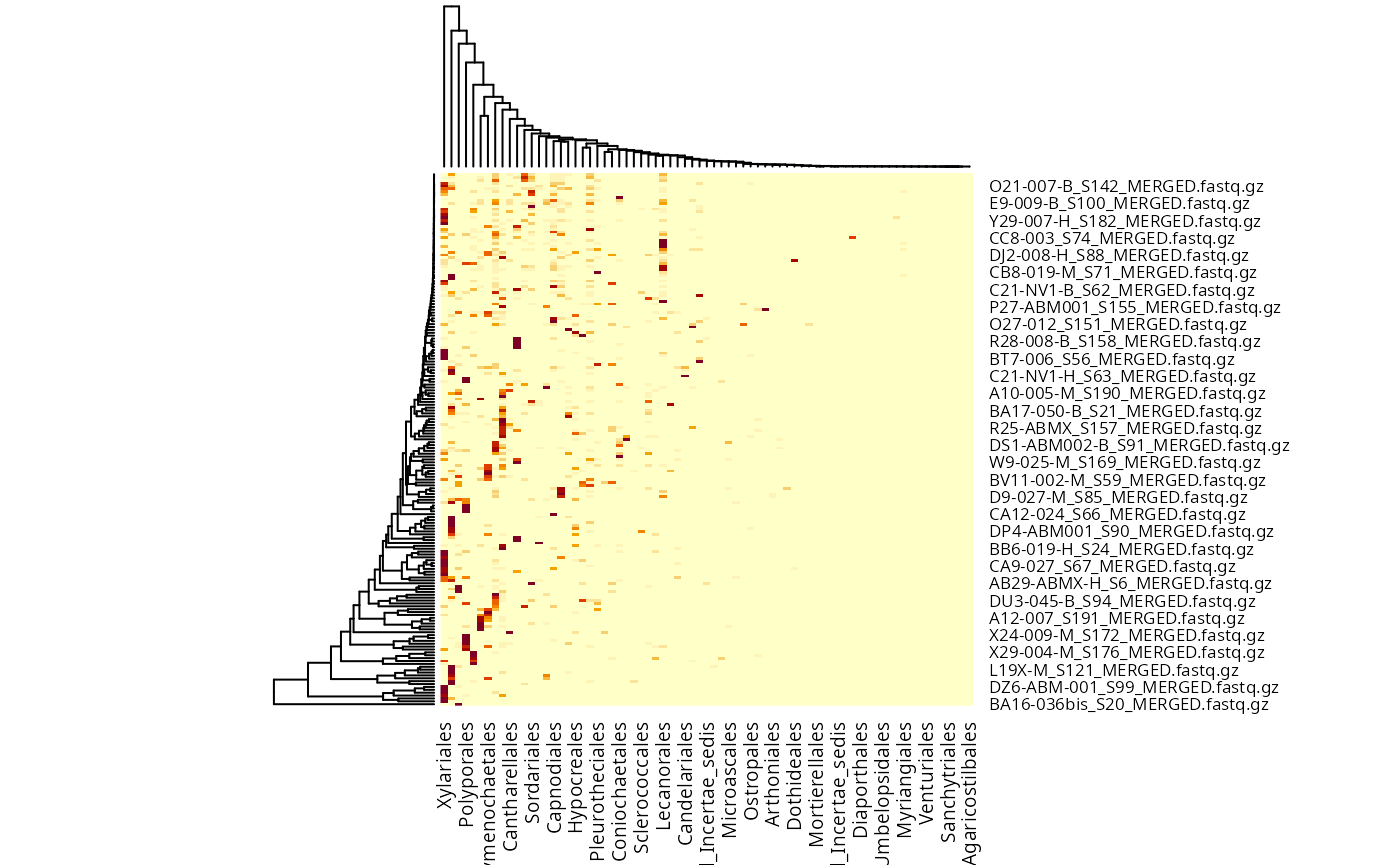
fungi_order <- tax_glom(data_fungi, taxrank = "Order")
taxa_names(fungi_order) <- fungi_order@tax_table[, "Order"]
heatmap(fungi_order@otu_table)