library(MiscMetabar)
data(data_fungi)Alpha diversity analysis
Hill number
Numerous metrics of diversity exist. Hill numbers 1 is a kind of general framework for alpha diversity index.
renyi_res <- vegan::renyi(data_fungi@otu_table)
head(renyi_res)
#> 0 0.25 0.5 1 2
#> A10-005-B_S188_MERGED.fastq.gz 4.204693 3.615323 3.044244 2.0754183 1.1561862
#> A10-005-H_S189_MERGED.fastq.gz 4.248495 3.150337 2.063712 0.9938545 0.6464533
#> A10-005-M_S190_MERGED.fastq.gz 3.988984 3.607773 3.308361 2.9304156 2.5781603
#> A12-007_S191_MERGED.fastq.gz 5.036953 4.423283 3.751946 2.6309045 1.8468497
#> A12-007-B_S2_MERGED.fastq.gz 3.850148 3.349461 2.874269 2.1881145 1.6533262
#> A15-004_S3_MERGED.fastq.gz 4.025352 3.890614 3.747555 3.4533352 2.9618789
#> 4 8 16 32
#> A10-005-B_S188_MERGED.fastq.gz 0.8007816 0.6864775 0.6407124 0.6200442
#> A10-005-H_S189_MERGED.fastq.gz 0.5146170 0.4469617 0.4172228 0.4037640
#> A10-005-M_S190_MERGED.fastq.gz 2.3022639 2.1015018 1.9744485 1.9110311
#> A12-007_S191_MERGED.fastq.gz 1.5920328 1.4947479 1.4421368 1.4109287
#> A12-007-B_S2_MERGED.fastq.gz 1.3979355 1.2774402 1.2106906 1.1733049
#> A15-004_S3_MERGED.fastq.gz 2.4997916 2.2378349 2.1045646 2.0377518
#> 64 Inf
#> A10-005-B_S188_MERGED.fastq.gz 0.6102022 0.6006678
#> A10-005-H_S189_MERGED.fastq.gz 0.3973550 0.3911464
#> A10-005-M_S190_MERGED.fastq.gz 1.8806976 1.8513117
#> A12-007_S191_MERGED.fastq.gz 1.3916365 1.3700776
#> A12-007-B_S2_MERGED.fastq.gz 1.1547034 1.1366612
#> A15-004_S3_MERGED.fastq.gz 2.0054156 1.9740810Test for difference in diversity (hill number)
One way to keep into account for difference in the number of sequences per samples is to use a Tukey test on a linear model with the square roots of the number of sequence as the first explanatory variable of the linear model 2.
p <- MiscMetabar::hill_pq(data_fungi, fact = "Height")
p$plot_Hill_0
#> NULL
p$plot_tuckey
#> NULLSee also the tutorial of the microbiome package for an alternative using the non-parametric Kolmogorov-Smirnov test for two-group comparisons when there are no relevant covariates.
Alpha diversity using package MicrobiotaProcess
library("MicrobiotaProcess")
clean_pq(subset_samples_pq(data_fungi, !is.na(data_fungi@sam_data$Height))) %>%
as.MPSE() %>%
mp_cal_alpha() %>%
mp_plot_alpha(.group = "Height")
#> Warning: `aes_string()` was deprecated in ggplot2 3.0.0.
#> ℹ Please use tidy evaluation idioms with `aes()`.
#> ℹ See also `vignette("ggplot2-in-packages")` for more information.
#> ℹ The deprecated feature was likely used in the MicrobiotaProcess package.
#> Please report the issue at
#> <https://github.com/YuLab-SMU/MicrobiotaProcess/issues>.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.
#> Warning in wilcox.test.default(c(4, 4, 5, 5, 4, 4, 3, 6, 3, 4, 3, 2, 3, :
#> cannot compute exact p-value with ties
#> Warning in wilcox.test.default(c(3, 4, 3, 5, 4, 6, 3, 5, 4, 4, 3, 3, 3, :
#> cannot compute exact p-value with ties
#> Warning in wilcox.test.default(c(3, 4, 3, 5, 4, 6, 3, 5, 4, 4, 3, 3, 3, :
#> cannot compute exact p-value with ties
#> Warning in wilcox.test.default(c(1.32966134885476, 1.242453324894,
#> 1.56071040904141, : cannot compute exact p-value with ties
#> Warning in wilcox.test.default(c(0.867563228481461, 1.32966134885476,
#> 0.867563228481461, : cannot compute exact p-value with ties
#> Warning in wilcox.test.default(c(0.867563228481461, 1.32966134885476,
#> 0.867563228481461, : cannot compute exact p-value with ties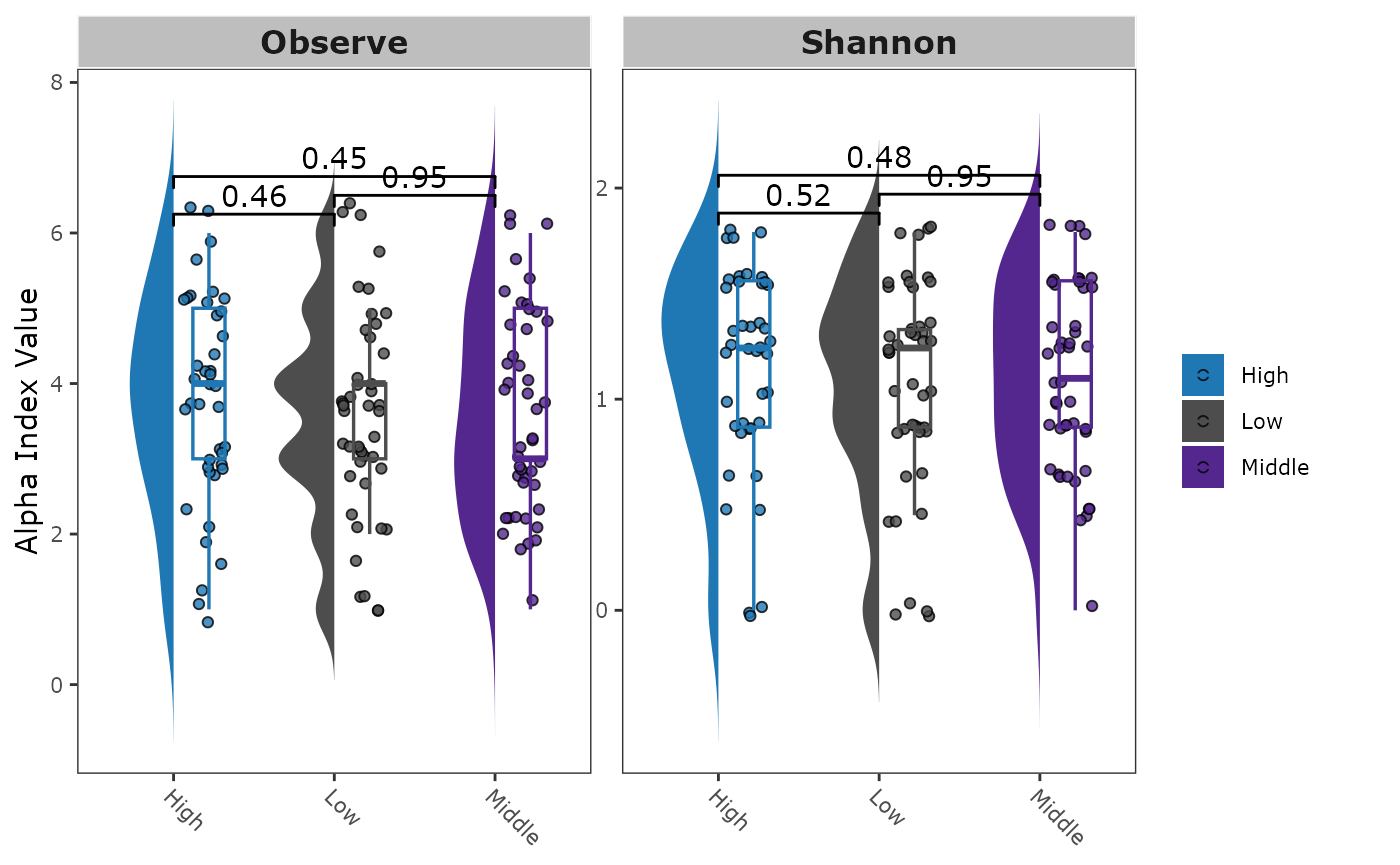
Durga package to represent and compute effect size of difference in alpha diversity
library("Durga")
psm <- psmelt_samples_pq(data_fungi)
d <- DurgaDiff(Hill_0 ~ Height, psm)
DurgaPlot(d)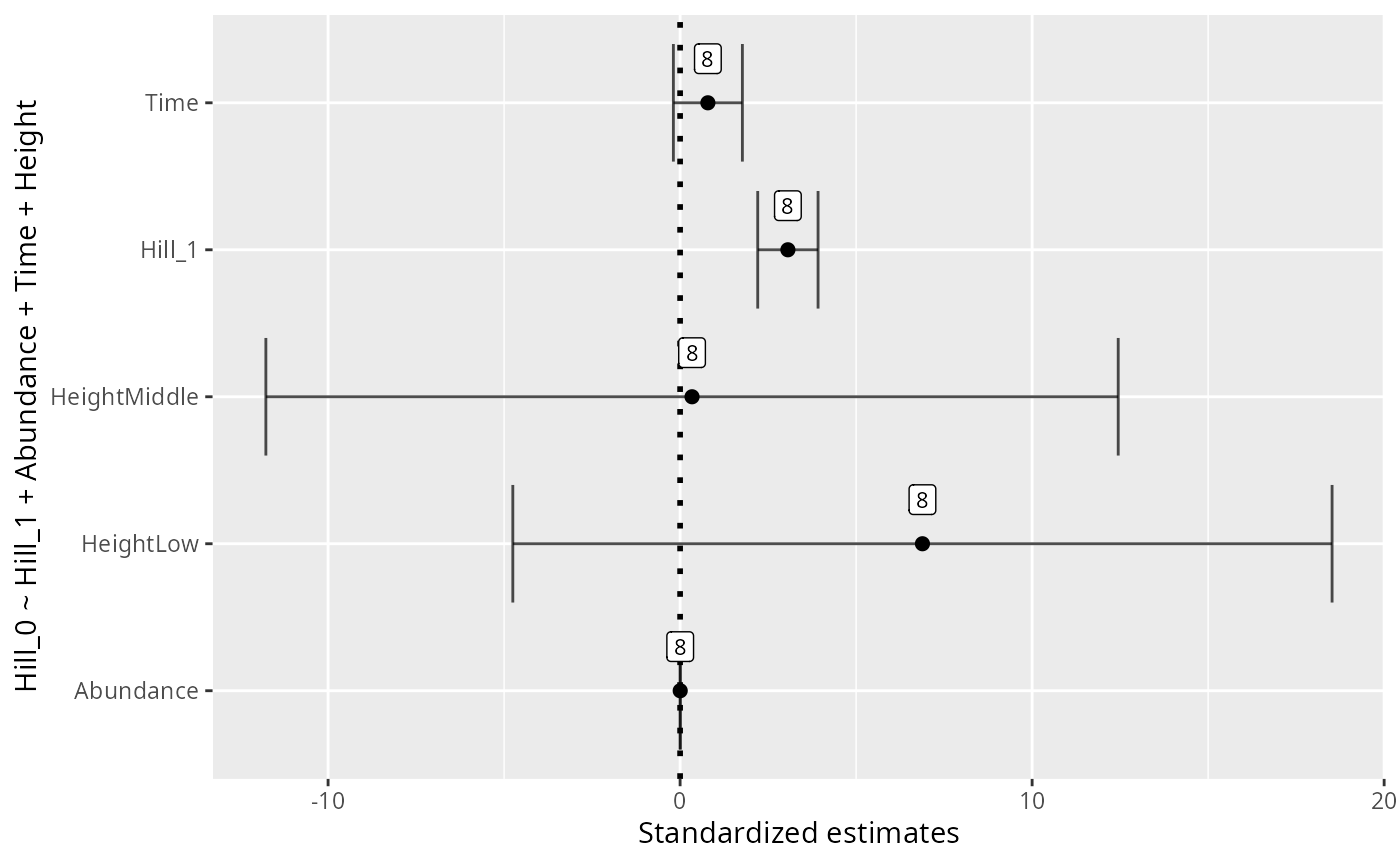
durga_pq <- function(physeq, formula, plot=FALSE) {
verify_pq(physeq)
psm <- psmelt_samples_pq(physeq)
res_durga <- DurgaDiff(formula, psm)
if(plot){
p <-DurgaPlot(res_durga)
invisible(p)
} else{
return(res_durga)
}
}
durga_pq(data_fungi, Hill_0 ~ Height, plot=TRUE)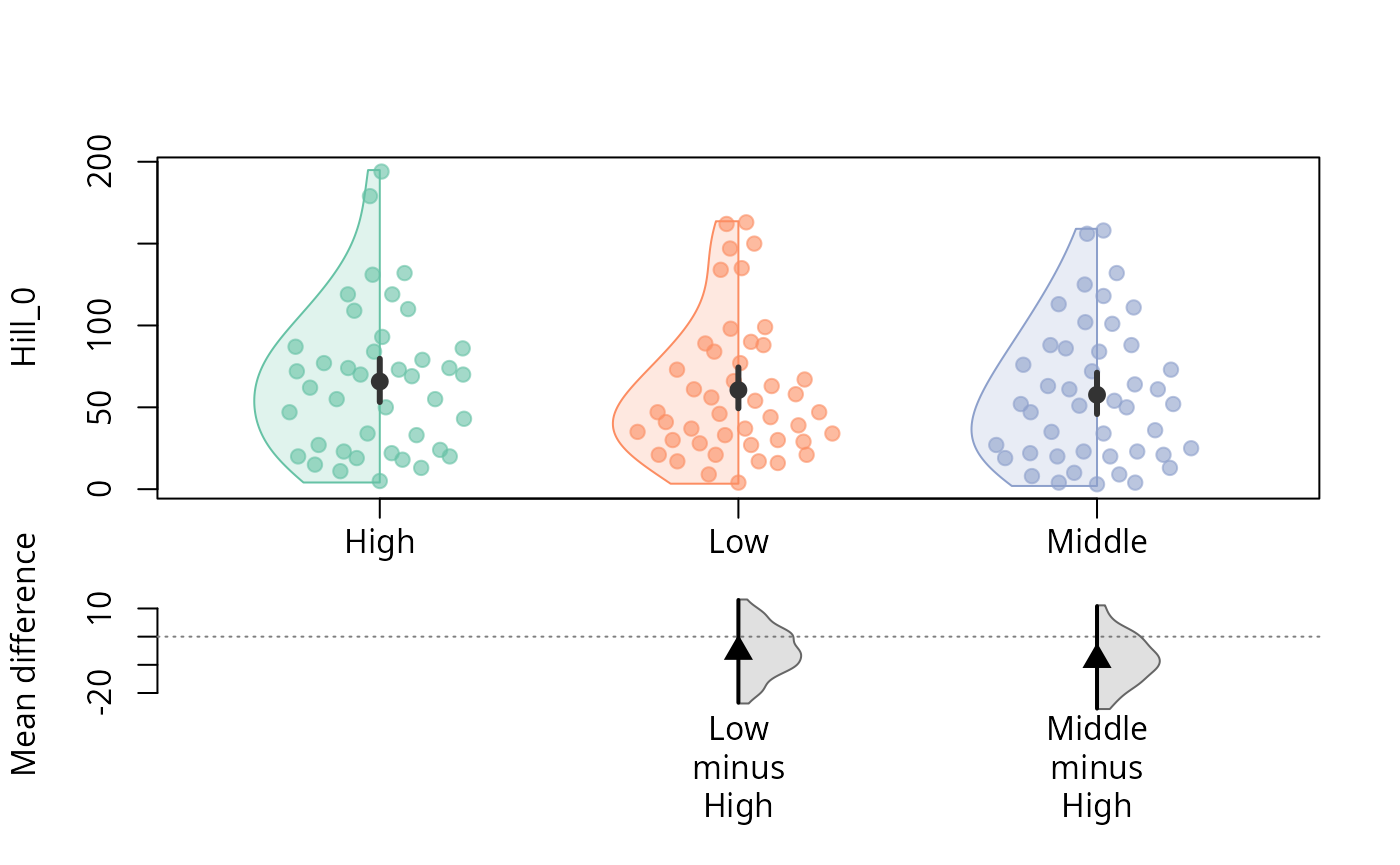
durga_pq(data_fungi, Hill_0 ~ Time + Height, plot=TRUE)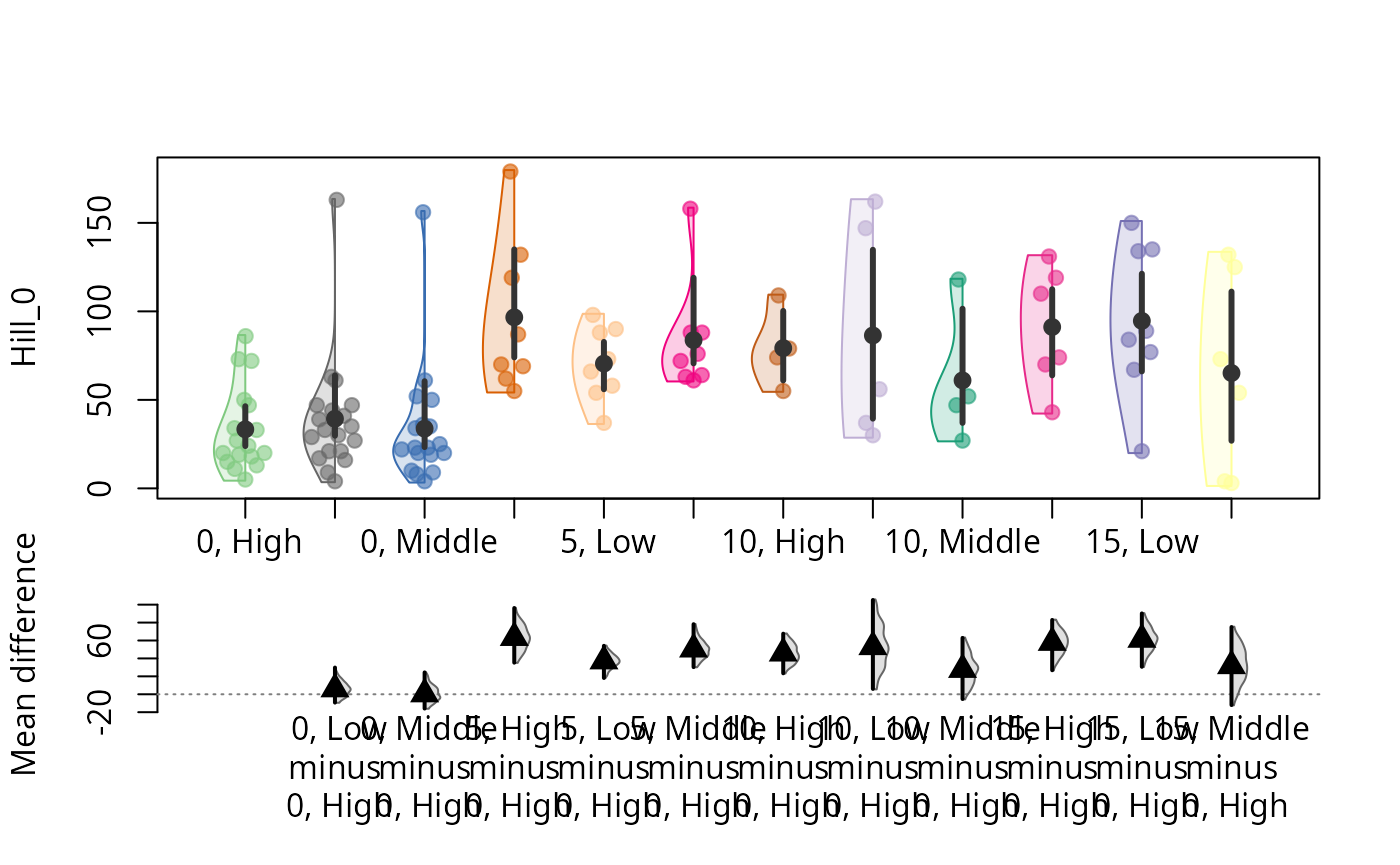
durga_pq(data_fungi, Hill_0 ~ Time==0, plot=TRUE)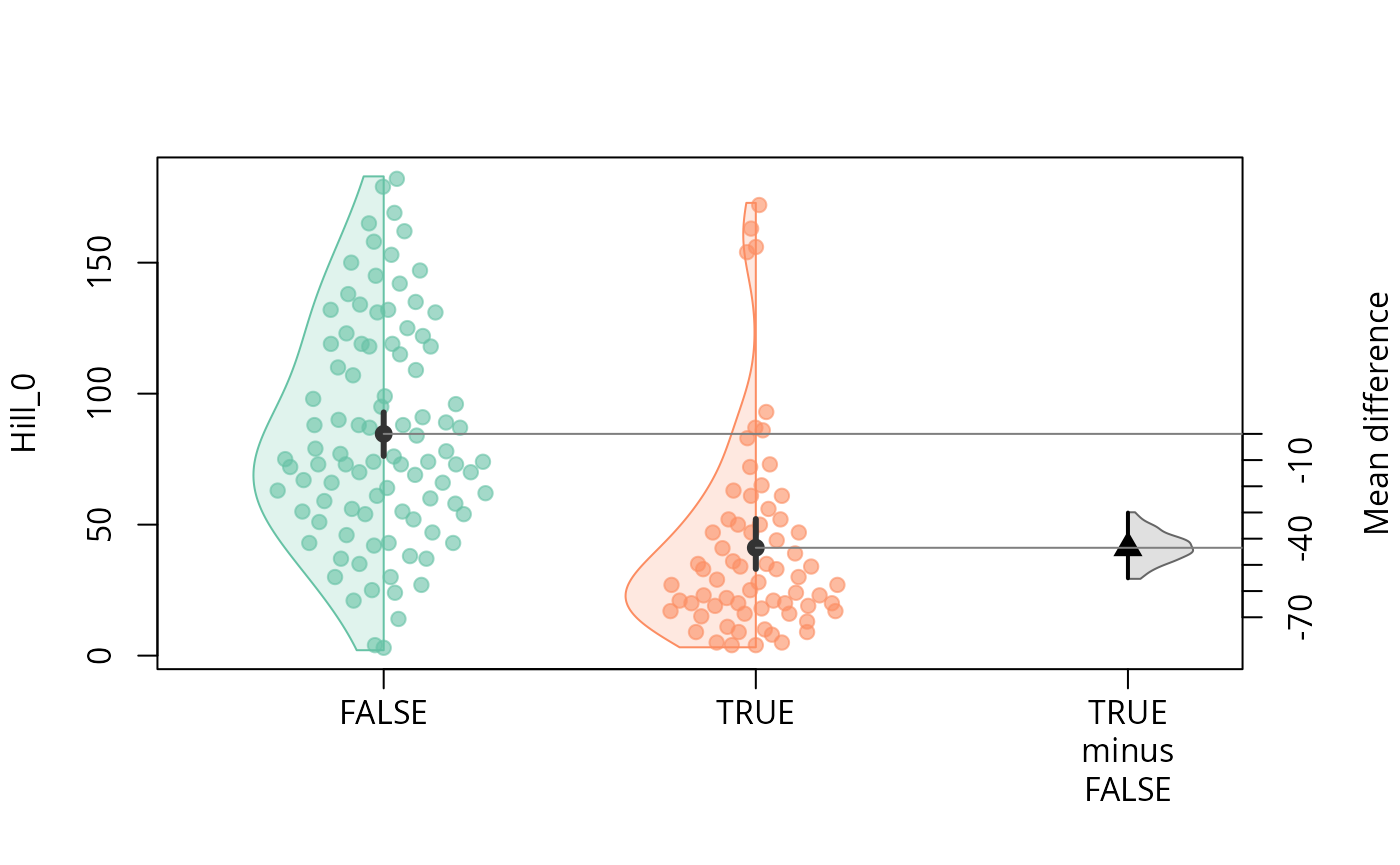
durga_pq(data_fungi, Hill_1 ~ Time==0, plot=TRUE)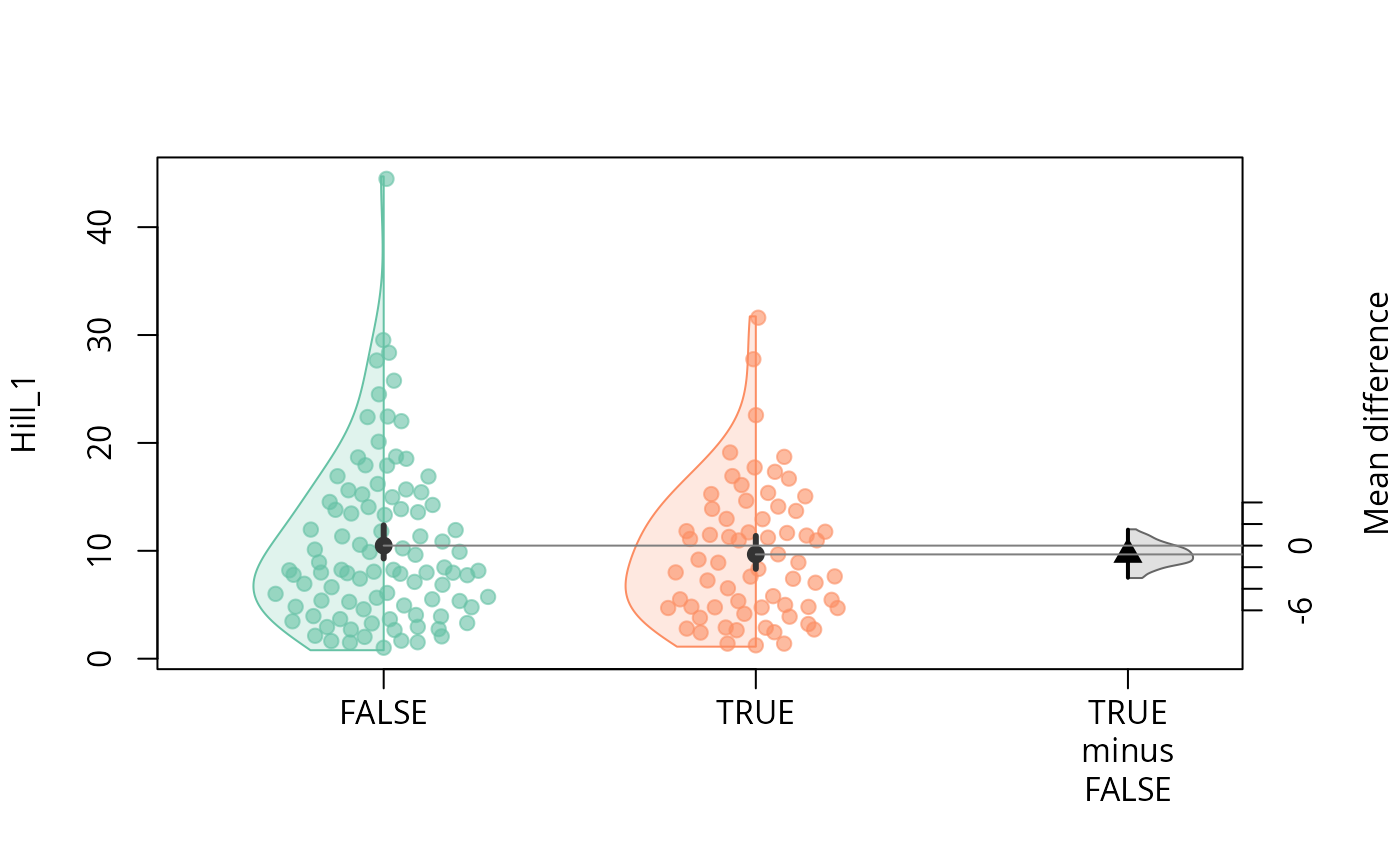
durga_pq(data_fungi, Hill_2 ~ Time==0, plot=TRUE)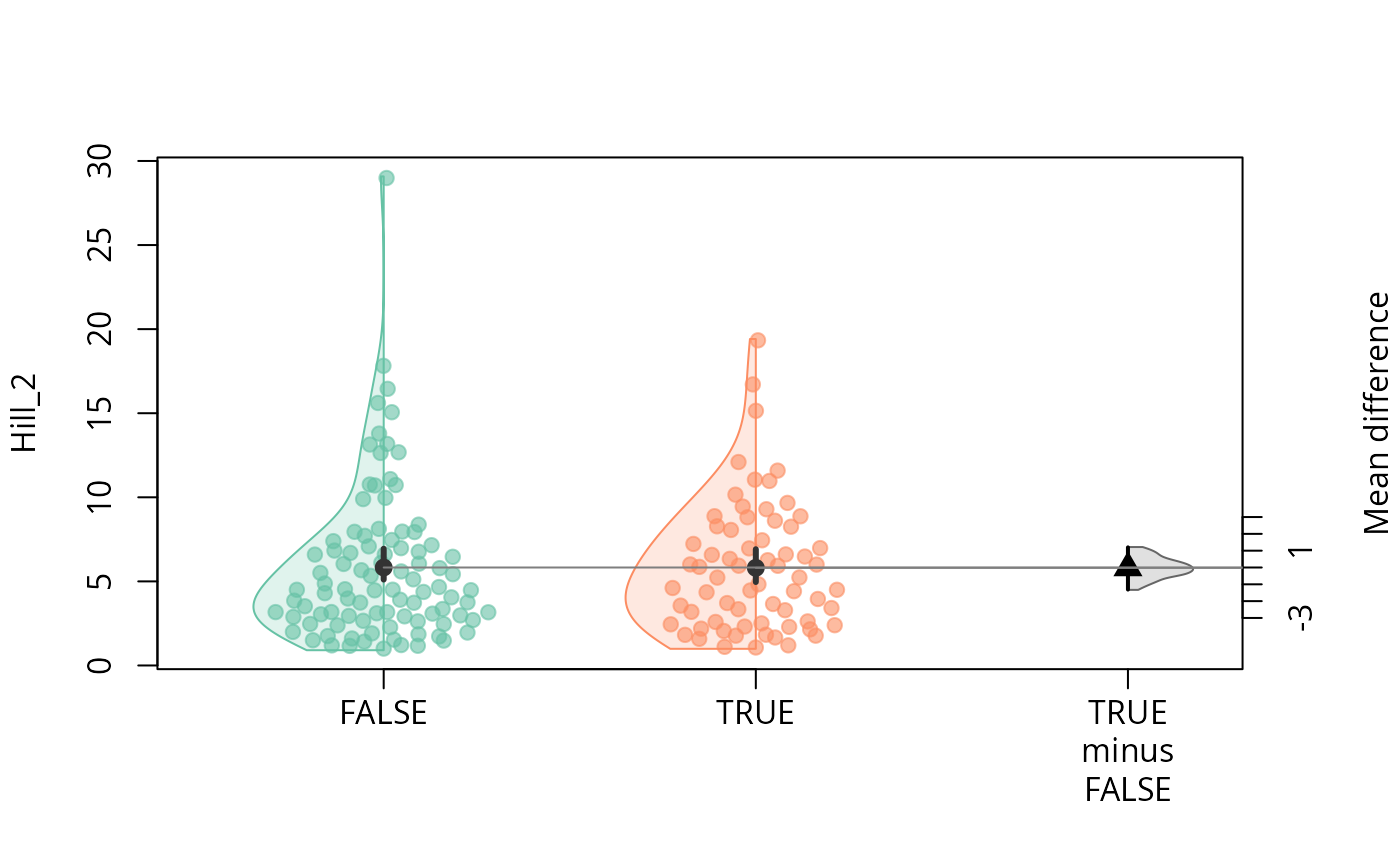
Effect of samples variables on alpha diversity using automated model selection and multimodel inference with (G)LMs
From the help of glmulti package :
glmulti finds what are the n best models (the confidence set of models) among all possible models (the candidate set, as specified by the user). Models are fitted with the specified fitting function (default is glm) and are ranked with the specified Information Criterion (default is aicc). The best models are found either through exhaustive screening of the candidates, or using a genetic algorithm, which allows very large candidate sets to be addressed. The output can be used for model selection, variable selection, and multimodel inference.
library("glmulti")
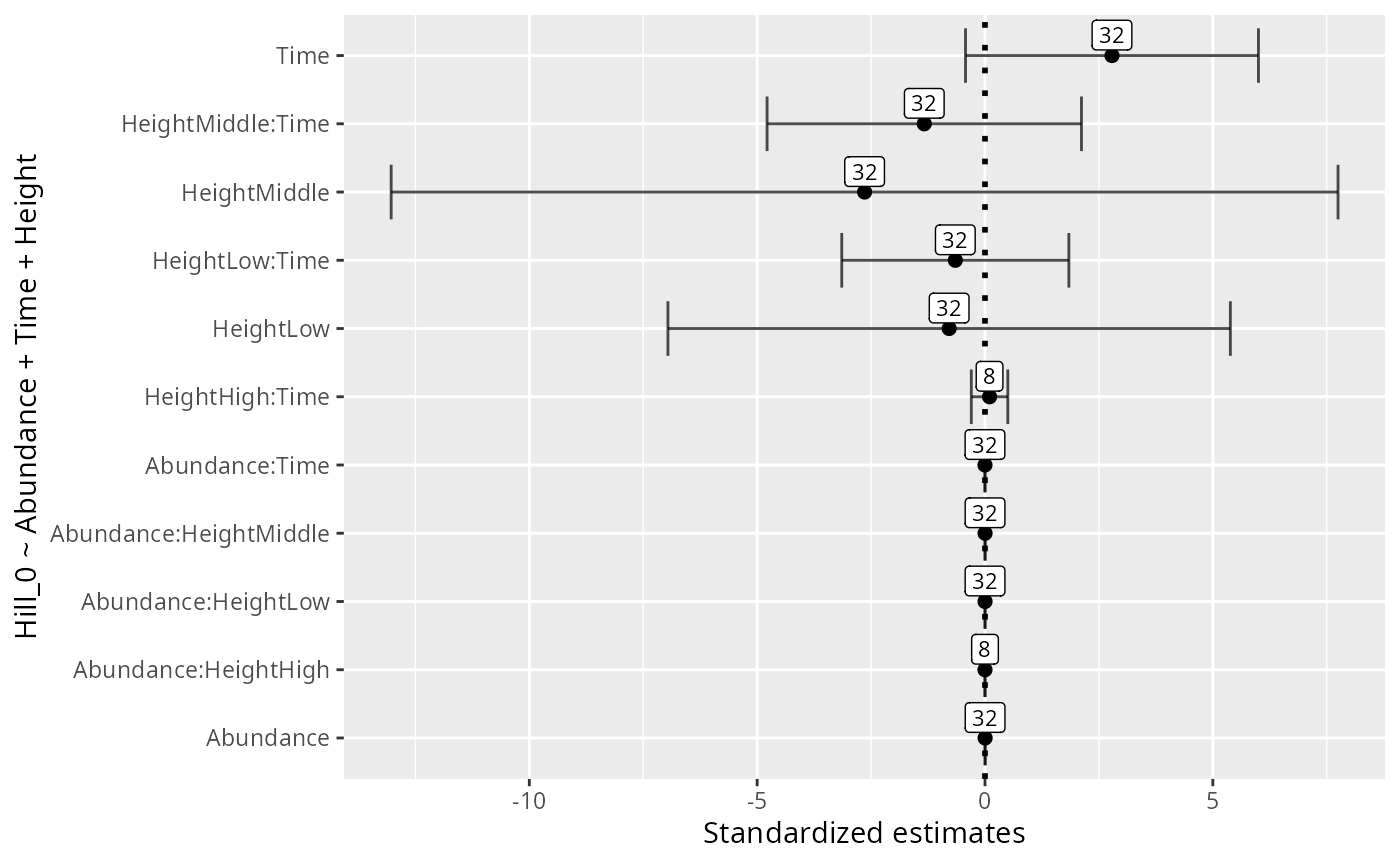
formula <- "Hill_0 ~ Hill_1 + Abundance + Time + Height"
res_glmulti <-
glmutli_pq(data_fungi, formula = formula, level = 1)
#> Initialization...
#> TASK: Exhaustive screening of candidate set.
#> Fitting...
#> Completed.
res_glmulti
#> estimates unconditional_interval nb_model importance
#> Hill_1 3.062117997 1.868174e-01 8 1
#> Abundance 0.002959644 8.478374e-08 8 1
#> Time 0.789091999 2.443263e-01 8 1
#> HeightLow 6.884340946 3.444196e+01 8 1
#> HeightMiddle 0.339123798 3.727962e+01 8 1
#> alpha variable
#> Hill_1 8.570200e-01 Hill_1
#> Abundance 5.773492e-04 Abundance
#> Time 9.800932e-01 Time
#> HeightLow 1.163660e+01 HeightLow
#> HeightMiddle 1.210648e+01 HeightMiddle
ggplot(data = res_glmulti, aes(x = estimates, y = variable)) +
geom_point(
size = 2,
alpha = 1,
show.legend = FALSE
) +
geom_vline(
xintercept = 0,
linetype = "dotted",
linewidth = 1
) +
geom_errorbar(
aes(xmin = estimates - alpha, xmax = estimates + alpha),
width = 0.8,
position = position_dodge(width = 0.8),
alpha = 0.7,
show.legend = FALSE
) +
geom_label(aes(label = nb_model), nudge_y = 0.3, size = 3) +
xlab("Standardized estimates") +
ylab(formula)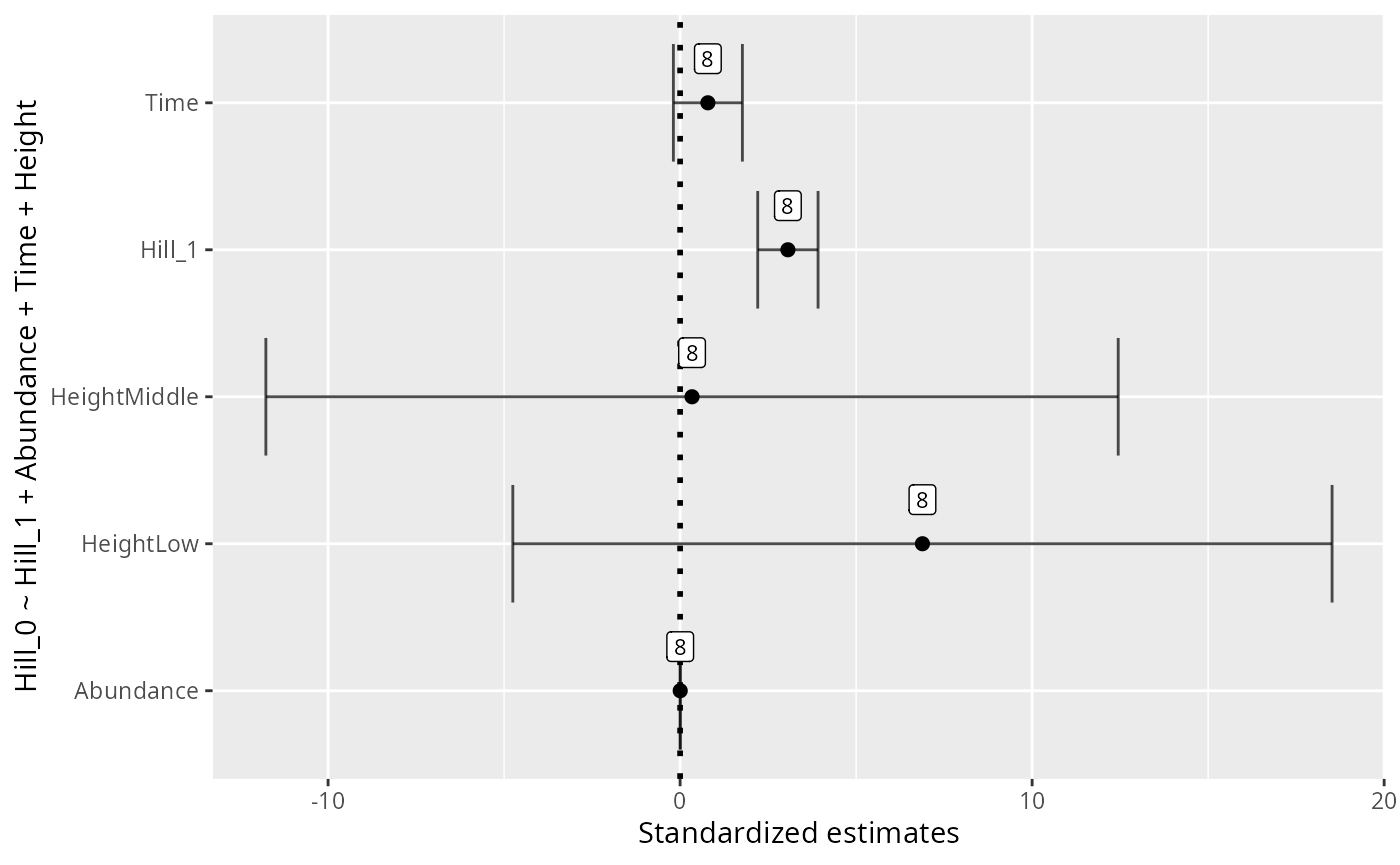
ggplot(data = res_glmulti, aes(
x = importance,
y = as.factor(variable),
fill = estimates
)) +
geom_bar(
stat = "identity",
show.legend = FALSE,
alpha = 0.8
) +
xlim(c(0, 1)) +
geom_label(aes(label = nb_model, x = 0.1),
size = 3,
fill = "white"
) +
scale_fill_viridis_b() +
xlab("Importance") +
ylab(formula)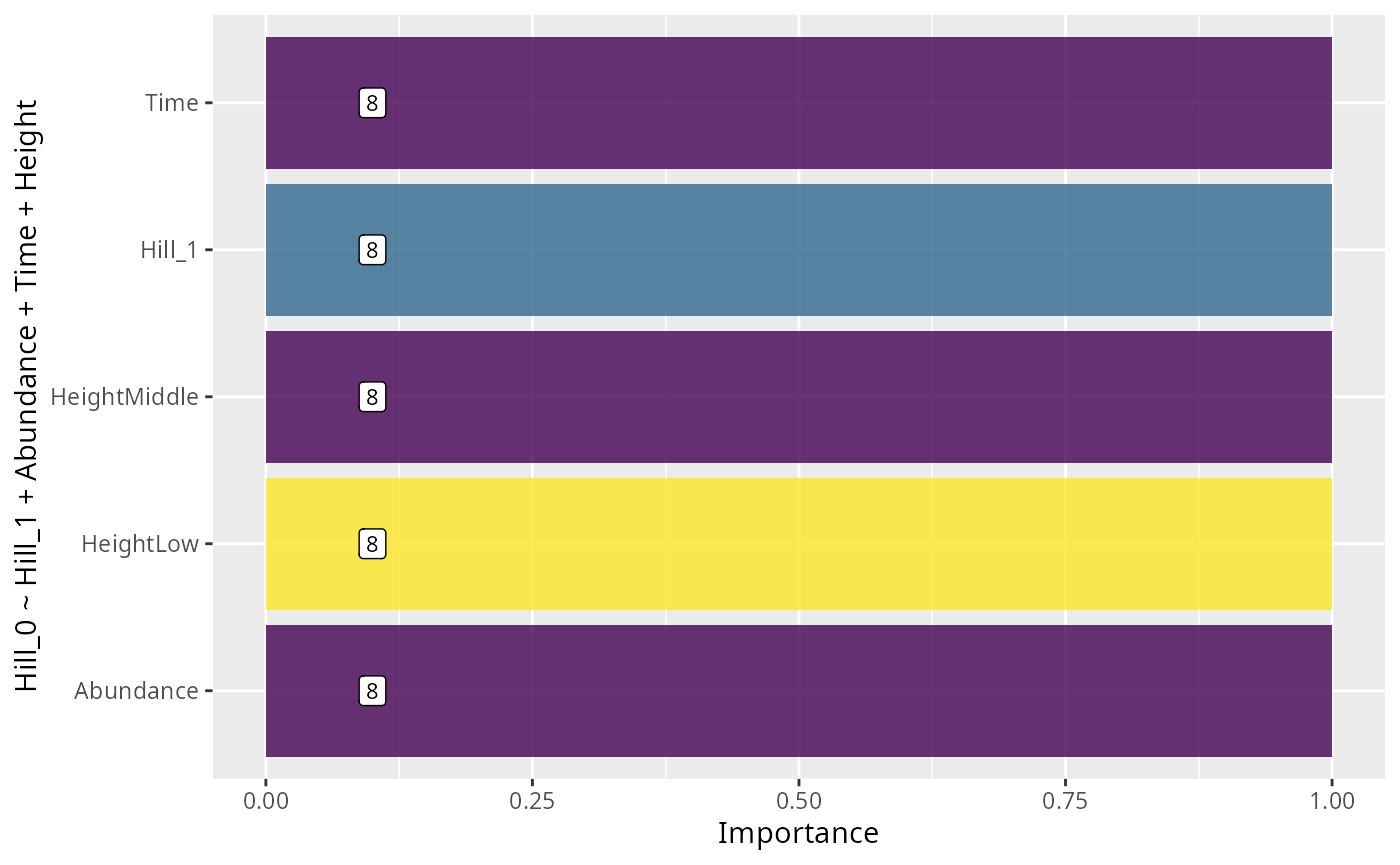
formula <- "Hill_0 ~ Abundance + Time + Height"
res_glmulti_interaction <-
glmutli_pq(data_fungi, formula = formula, level = 2)
#> Initialization...
#> TASK: Exhaustive screening of candidate set.
#> Fitting...
#>
#> After 50 models:
#> Best model: Hill_0~1+Abundance+Time+Time:Abundance+Height:Abundance+Height:Time
#> Crit= 1069.11608982306
#> Mean crit= 1218.19009955263
#> Completed.
res_glmulti_interaction
#> estimates unconditional_interval nb_model importance
#> HeightHigh:Time 0.1004073616 4.167750e-02 8 0.04216251
#> Abundance:HeightHigh 0.0001609310 8.984023e-08 8 0.09020701
#> HeightLow -0.7865687564 9.769200e+00 32 0.24714664
#> HeightMiddle -2.6419930721 2.789953e+01 32 0.24714664
#> HeightLow:Time -0.6511123699 1.599292e+00 32 0.55051517
#> HeightMiddle:Time -1.3322473025 3.078720e+00 32 0.55051517
#> Abundance:Time -0.0001068559 4.586032e-09 32 0.81587143
#> Abundance:HeightLow 0.0011137659 7.957713e-07 32 0.86967993
#> Abundance:HeightMiddle 0.0017155970 1.245718e-06 32 0.86967993
#> Abundance 0.0024839088 8.790126e-07 32 0.90902176
#> Time 2.7869220741 2.663548e+00 32 0.92512111
#> alpha variable
#> HeightHigh:Time 4.006348e-01 HeightHigh:Time
#> Abundance:HeightHigh 5.877776e-04 Abundance:HeightHigh
#> HeightLow 6.171172e+00 HeightLow
#> HeightMiddle 1.038989e+01 HeightMiddle
#> HeightLow:Time 2.491898e+00 HeightLow:Time
#> HeightMiddle:Time 3.449710e+00 HeightMiddle:Time
#> Abundance:Time 1.335247e-04 Abundance:Time
#> Abundance:HeightLow 1.759641e-03 Abundance:HeightLow
#> Abundance:HeightMiddle 2.198334e-03 Abundance:HeightMiddle
#> Abundance 1.847287e-03 Abundance
#> Time 3.214276e+00 Time
ggplot(data = res_glmulti_interaction, aes(x = estimates, y = variable)) +
geom_point(
size = 2,
alpha = 1,
show.legend = FALSE
) +
geom_vline(
xintercept = 0,
linetype = "dotted",
linewidth = 1
) +
geom_errorbar(
aes(xmin = estimates - alpha, xmax = estimates + alpha),
width = 0.8,
position = position_dodge(width = 0.8),
alpha = 0.7,
show.legend = FALSE
) +
geom_label(aes(label = nb_model), nudge_y = 0.3, size = 3) +
xlab("Standardized estimates") +
ylab(formula)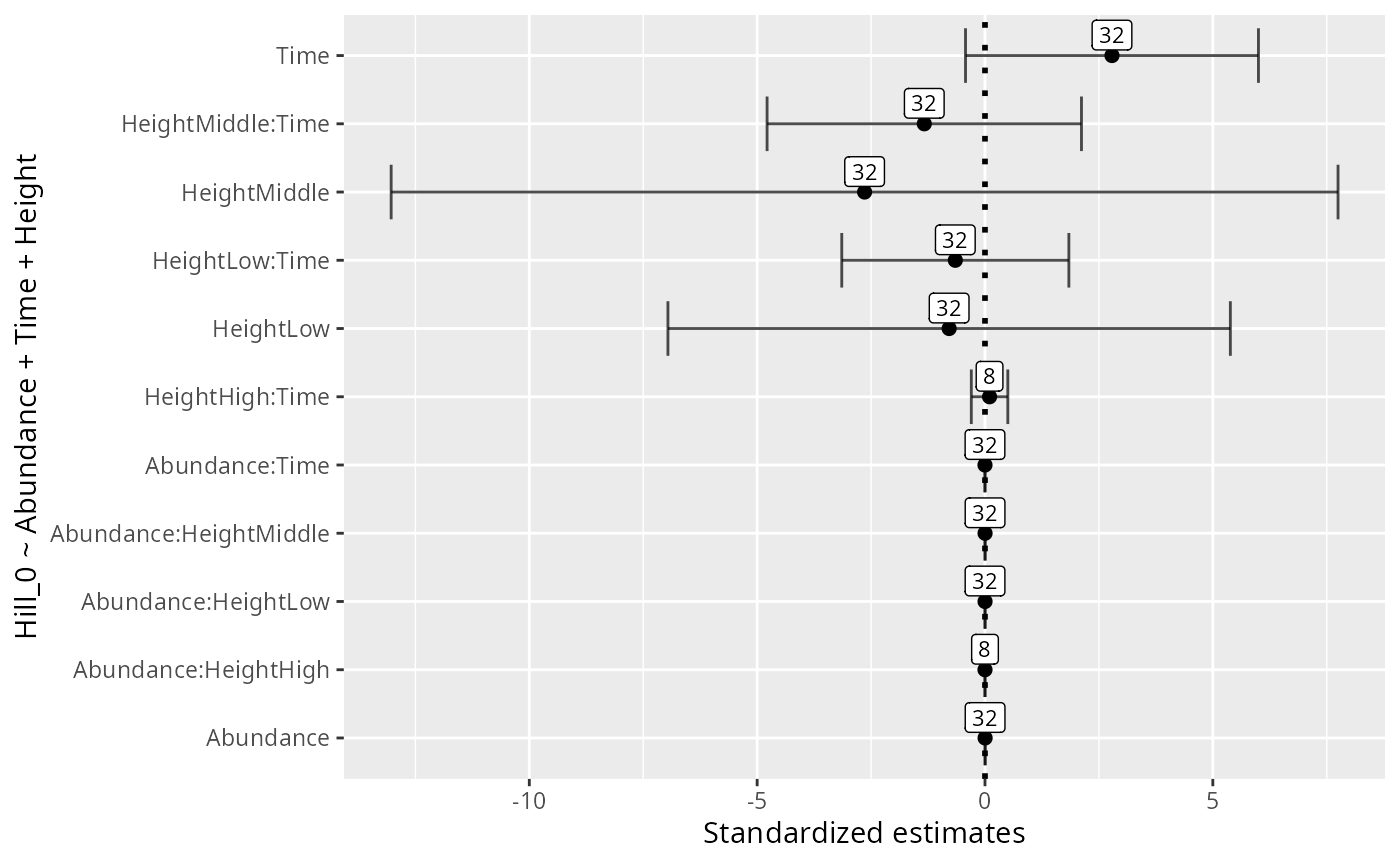
ggplot(data = res_glmulti_interaction, aes(
x = importance,
y = as.factor(variable),
fill = estimates
)) +
geom_bar(
stat = "identity",
show.legend = FALSE,
alpha = 0.8
) +
xlim(c(0, 1)) +
geom_label(aes(label = nb_model, x = 0.1),
size = 3,
fill = "white"
) +
scale_fill_viridis_b() +
xlab("Importance") +
ylab(formula)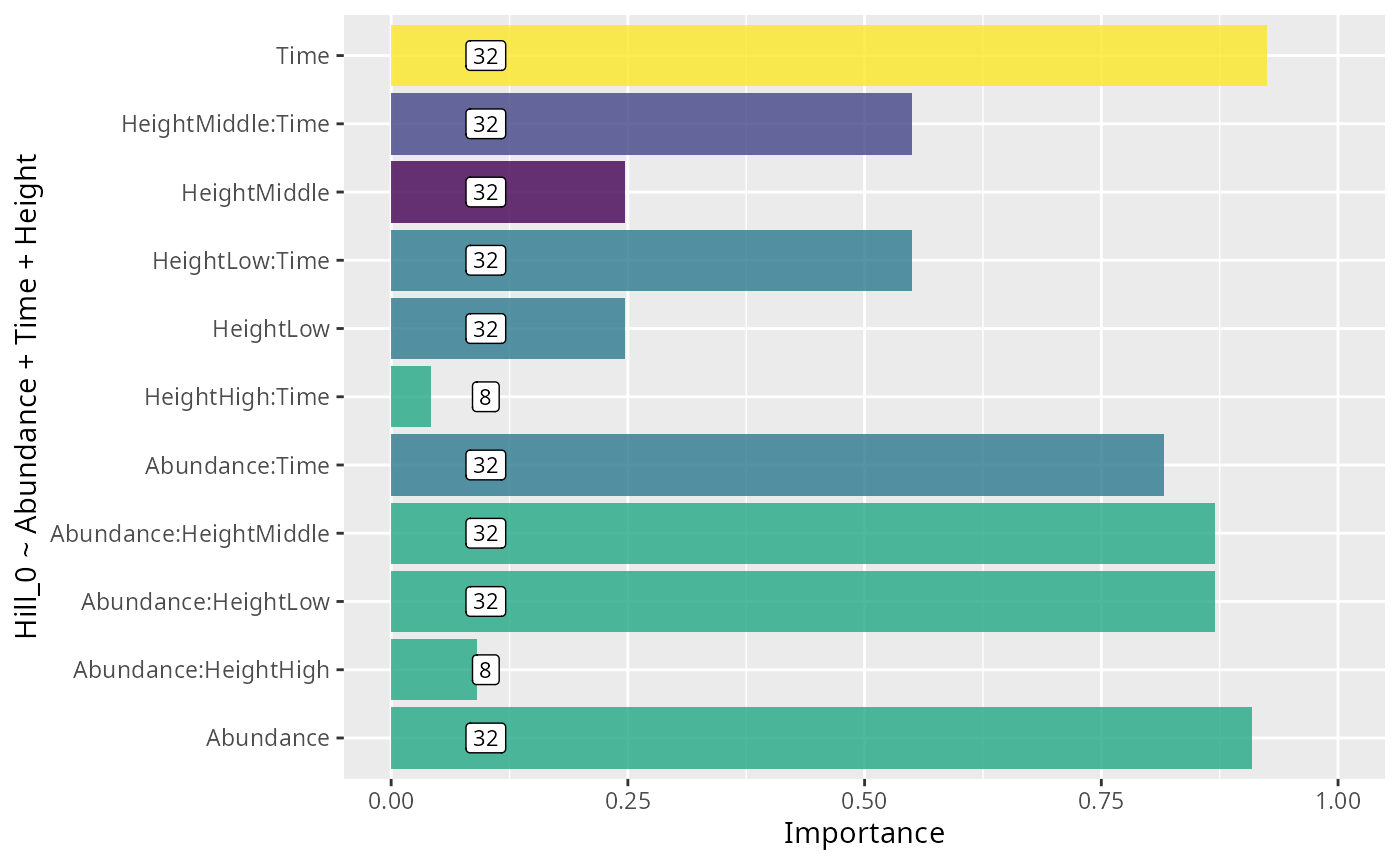
Session information
sessionInfo()
#> R version 4.5.2 (2025-10-31)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Kali GNU/Linux Rolling
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.29.so; LAPACK version 3.12.0
#>
#> locale:
#> [1] LC_CTYPE=fr_FR.UTF-8 LC_NUMERIC=C
#> [3] LC_TIME=fr_FR.UTF-8 LC_COLLATE=fr_FR.UTF-8
#> [5] LC_MONETARY=fr_FR.UTF-8 LC_MESSAGES=fr_FR.UTF-8
#> [7] LC_PAPER=fr_FR.UTF-8 LC_NAME=fr_FR.UTF-8
#> [9] LC_ADDRESS=fr_FR.UTF-8 LC_TELEPHONE=fr_FR.UTF-8
#> [11] LC_MEASUREMENT=fr_FR.UTF-8 LC_IDENTIFICATION=fr_FR.UTF-8
#>
#> time zone: Europe/Paris
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] glmulti_1.0.8 leaps_3.2 rJava_1.0-11
#> [4] Durga_2.1.0 MicrobiotaProcess_1.22.0 MiscMetabar_0.14.5
#> [7] purrr_1.2.0 dplyr_1.1.4 dada2_1.38.0
#> [10] Rcpp_1.1.0 ggplot2_4.0.1 phyloseq_1.54.0
#>
#> loaded via a namespace (and not attached):
#> [1] libcoin_1.0-10 RColorBrewer_1.1-3
#> [3] jsonlite_2.0.0 magrittr_2.0.4
#> [5] TH.data_1.1-5 modeltools_0.2-24
#> [7] farver_2.1.2 rmarkdown_2.30
#> [9] fs_1.6.6 ragg_1.5.0
#> [11] vctrs_0.6.5 multtest_2.66.0
#> [13] Rsamtools_2.26.0 ggtree_4.0.1
#> [15] htmltools_0.5.9 S4Arrays_1.10.1
#> [17] Rhdf5lib_1.32.0 gridGraphics_0.5-1
#> [19] SparseArray_1.10.7 rhdf5_2.54.1
#> [21] sass_0.4.10 bslib_0.9.0
#> [23] htmlwidgets_1.6.4 desc_1.4.3
#> [25] plyr_1.8.9 sandwich_3.1-1
#> [27] zoo_1.8-15 cachem_1.1.0
#> [29] GenomicAlignments_1.46.0 igraph_2.2.1
#> [31] lifecycle_1.0.4 iterators_1.0.14
#> [33] pkgconfig_2.0.3 Matrix_1.7-4
#> [35] R6_2.6.1 fastmap_1.2.0
#> [37] MatrixGenerics_1.22.0 digest_0.6.39
#> [39] aplot_0.2.9 ggnewscale_0.5.2
#> [41] ShortRead_1.68.0 patchwork_1.3.2
#> [43] S4Vectors_0.48.0 textshaping_1.0.4
#> [45] GenomicRanges_1.62.1 hwriter_1.3.2.1
#> [47] vegan_2.7-2 labeling_0.4.3
#> [49] abind_1.4-8 mgcv_1.9-4
#> [51] compiler_4.5.2 fontquiver_0.2.1
#> [53] withr_3.0.2 S7_0.2.1
#> [55] BiocParallel_1.44.0 ggsignif_0.6.4
#> [57] MASS_7.3-65 rappdirs_0.3.3
#> [59] DelayedArray_0.36.0 biomformat_1.38.0
#> [61] permute_0.9-8 tools_4.5.2
#> [63] vipor_0.4.7 ape_5.8-1
#> [65] glue_1.8.0 nlme_3.1-168
#> [67] rhdf5filters_1.22.0 grid_4.5.2
#> [69] cluster_2.1.8.1 reshape2_1.4.5
#> [71] ade4_1.7-23 generics_0.1.4
#> [73] gtable_0.3.6 tidyr_1.3.1
#> [75] data.table_1.17.8 coin_1.4-3
#> [77] XVector_0.50.0 BiocGenerics_0.56.0
#> [79] ggrepel_0.9.6 foreach_1.5.2
#> [81] pillar_1.11.1 stringr_1.6.0
#> [83] yulab.utils_0.2.3 splines_4.5.2
#> [85] treeio_1.34.0 lattice_0.22-7
#> [87] survival_3.8-3 deldir_2.0-4
#> [89] tidyselect_1.2.1 fontLiberation_0.1.0
#> [91] Biostrings_2.78.0 knitr_1.50
#> [93] fontBitstreamVera_0.1.1 gridExtra_2.3
#> [95] IRanges_2.44.0 Seqinfo_1.0.0
#> [97] SummarizedExperiment_1.40.0 ggtreeExtra_1.20.0
#> [99] stats4_4.5.2 xfun_0.55
#> [101] Biobase_2.70.0 matrixStats_1.5.0
#> [103] stringi_1.8.7 boot_1.3-32
#> [105] lazyeval_0.2.2 ggfun_0.2.0
#> [107] yaml_2.3.12 evaluate_1.0.5
#> [109] codetools_0.2-20 cigarillo_1.0.0
#> [111] interp_1.1-6 gdtools_0.4.4
#> [113] tibble_3.3.0 ggplotify_0.1.3
#> [115] cli_3.6.5 RcppParallel_5.1.11-1
#> [117] systemfonts_1.3.1 jquerylib_0.1.4
#> [119] png_0.1-8 parallel_4.5.2
#> [121] ggh4x_0.3.1 pkgdown_2.2.0
#> [123] latticeExtra_0.6-31 jpeg_0.1-11
#> [125] bitops_1.0-9 ggstar_1.0.6
#> [127] pwalign_1.6.0 viridisLite_0.4.2
#> [129] mvtnorm_1.3-3 tidytree_0.4.6
#> [131] ggiraph_0.9.2 scales_1.4.0
#> [133] crayon_1.5.3 rlang_1.1.6
#> [135] multcomp_1.4-29