Run Aldex on a phyloseq object
Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- bifactor
(required) The name of a column present in the
@sam_dataslot of the physeq object. Must be a character vector or a factor.- modalities
(default NULL) A vector of modalities to keep in the analysis. If NULL, all modalities present in bifactor are kept. Note that only two modalities are allowed. @param gamma (default 0.5) The value of the Dirichlet Monte-Carlo sampling parameter. @param ... Additional arguments passed on to
ALDEx2::aldex()
Value
The result of ALDEx2::aldex()
Details
It is a wrapper of the ALDEx2::aldex() function with default gamma=0.5.
Examples
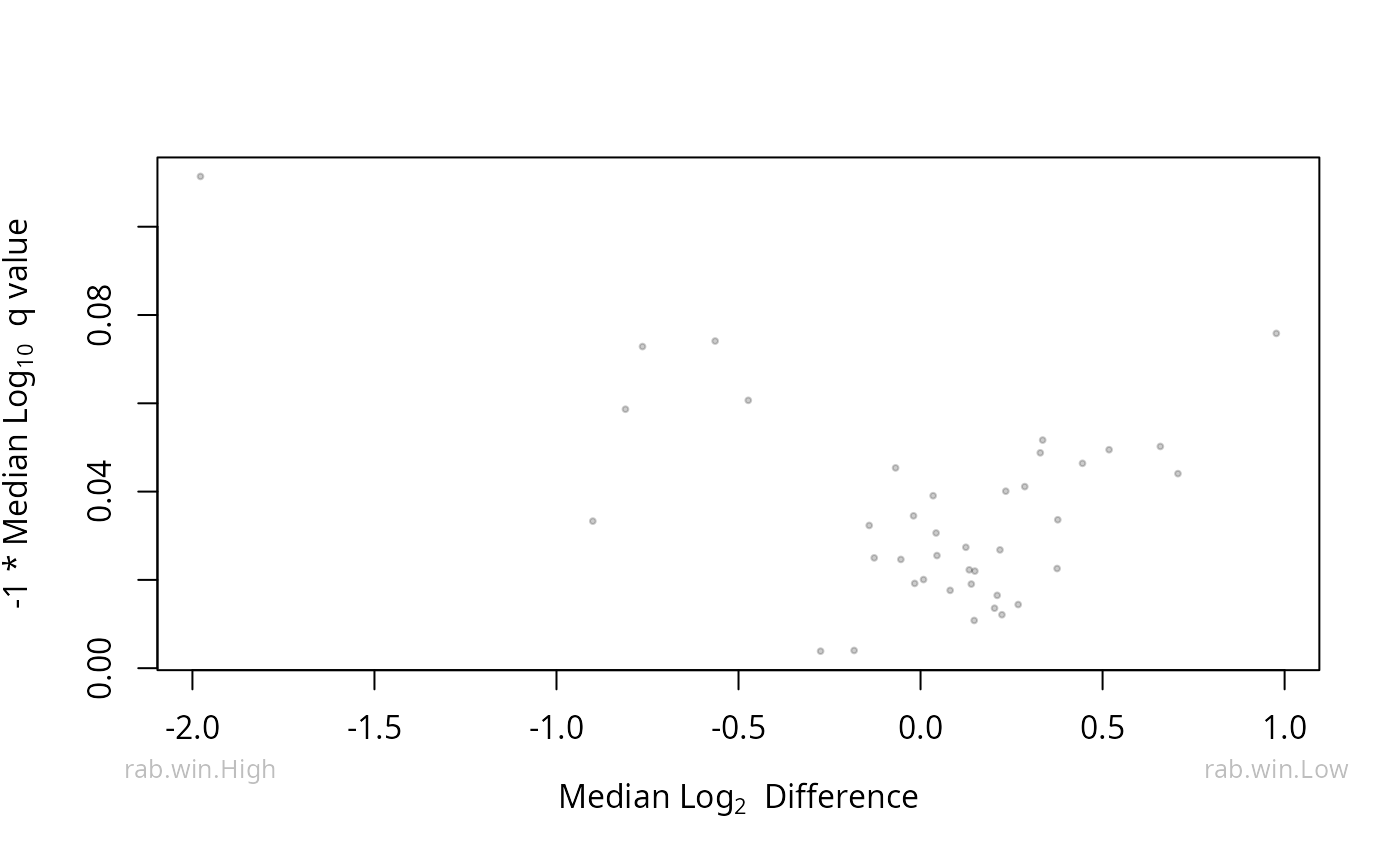
res_aldex <- aldex_pq(data_fungi_mini,
bifactor = "Height",
modalities = c("Low", "High")
)
#> aldex.clr: generating Monte-Carlo instances and clr values
#> conditions vector supplied
#> operating in serial mode
#> aldex.scaleSim: adjusting samples to reflect scale uncertainty.
#> aldex.ttest: doing t-test
#> aldex.effect: calculating effect sizes
ALDEx2::aldex.plot(res_aldex, type = "volcano")