Test if the mean number of sequences by samples is link to the modality of a factor
Source:R/dada_phyloseq.R
are_modality_even_depth.RdThe aim of this function is to provide a warnings if samples depth significantly
vary among the modalities of a factor present in the sam_data slot.
This function apply a Kruskal-Wallis rank sum test to the number of sequences
per samples in function of the factor fact.
Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- fact
(required) Name of the factor to cluster samples by modalities. Need to be in
physeq@sam_data.- boxplot
(logical) Do you want to plot boxplot?
Examples
are_modality_even_depth(data_fungi_mini, "Time")$p.value
#> [1] 0.0006936505
are_modality_even_depth(rarefy_even_depth(data_fungi_mini), "Time")$p.value
#> You set `rngseed` to FALSE. Make sure you've set & recorded
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> ...
#> 6OTUs were removed because they are no longer
#> present in any sample after random subsampling
#> ...
#> All modality were undoubtedly rarefy in the physeq object.
#> [1] 1
are_modality_even_depth(data_fungi_mini, "Height", boxplot = TRUE)
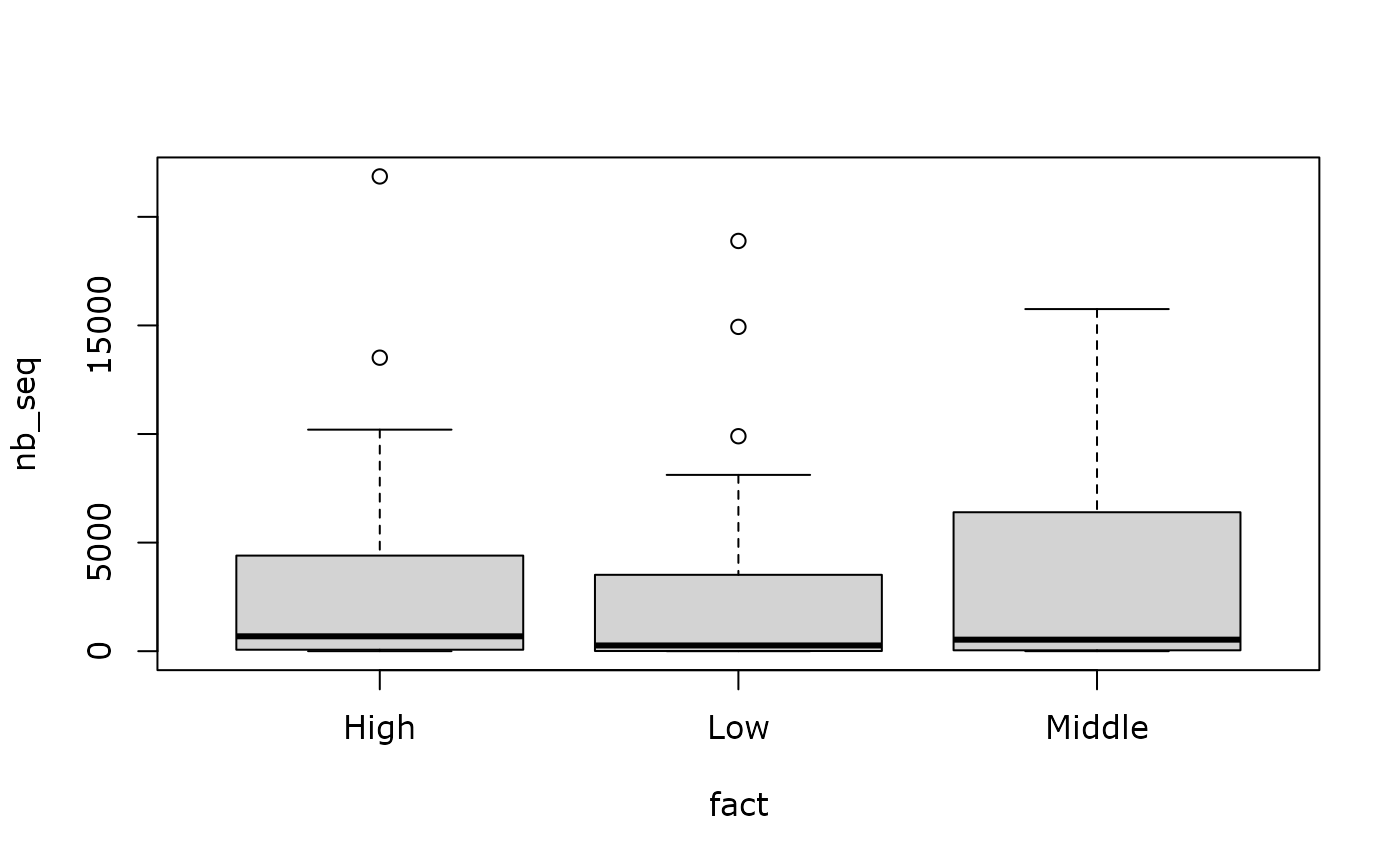 #>
#> Kruskal-Wallis rank sum test
#>
#> data: nb_seq by fact
#> Kruskal-Wallis chi-squared = 1.1143, df = 2, p-value = 0.5728
#>
#>
#> Kruskal-Wallis rank sum test
#>
#> data: nb_seq by fact
#> Kruskal-Wallis chi-squared = 1.1143, df = 2, p-value = 0.5728
#>
