Basically a wrapper of ggalluvial package
Usage
ggaluv_pq(
physeq,
taxa_ranks = c("Phylum", "Class", "Order", "Family"),
wrap_factor = NULL,
by_sample = FALSE,
rarefy_by_sample = FALSE,
rngseed = FALSE,
verbose = TRUE,
fact = NULL,
type = "nb_seq",
width = 1.2,
min.size = 3,
na_remove = FALSE,
use_ggfittext = FALSE,
use_geom_label = FALSE,
size_lab = 2,
...
)Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- taxa_ranks
A vector of taxonomic ranks. For examples c("Family","Genus"). If taxa ranks is not set (default value = c("Phylum", "Class", "Order", "Family")).
- wrap_factor
A name to determine which samples to merge using
merge_samples2()function. Need to be inphyseq@sam_data. Need to be use when you want to wrap by factor the final plot with the number of taxa (type="nb_taxa")- by_sample
(logical) If FALSE (default), sample information is not taking into account, so the taxonomy is studied globally. If fact is not NULL, by_sample is automatically set to TRUE.
- rarefy_by_sample
(logical, default FALSE) If TRUE, rarefy samples using
phyloseq::rarefy_even_depth()function.- rngseed
(Optional). A single integer value passed to
phyloseq::rarefy_even_depth(), which is used to fix a seed for reproducibly random number generation (in this case, reproducibly random subsampling). If set to FALSE, then no fiddling with the RNG seed is performed, and it is up to the user to appropriately call set.seed beforehand to achieve reproducible results. Default is FALSE.- verbose
(logical). If TRUE, print additional information.
- fact
(required) Name of the factor in
physeq@sam_dataused to plot the last column- type
If "nb_seq" (default), the number of sequences is used in plot. If "nb_taxa", the number of ASV is plotted.
- width
(passed on to
ggalluvial::geom_flow()) the width of each stratum, as a proportion of the distance between axes. Defaults to 1/3.- min.size
(passed on to
ggfittext::geom_fit_text()) Minimum font size, in points. Text that would need to be shrunk below this size to fit the box will be hidden. Defaults to 4 pt.- na_remove
(logical, default FALSE) If set to TRUE, remove samples with NA in the variables set in formula.
- use_ggfittext
(logical, default FALSE) Do we use ggfittext to plot labels?
- use_geom_label
(logical, default FALSE) Do we use geom_label to plot labels?
- size_lab
Size for label if use_ggfittext is FALSE
- ...
Additional arguments passed on to
ggalluvial::geom_flow()function.
Details
This function is mainly a wrapper of the work of others.
Please make a reference to ggalluvial package if you
use this function.
When you want to add text to the plot, this function requires
ggalluvial to be loaded with before use (library(ggalluvial)).
Examples
if (requireNamespace("ggalluvial")) {
ggaluv_pq(data_fungi_mini)
}
#> Loading required namespace: ggalluvial
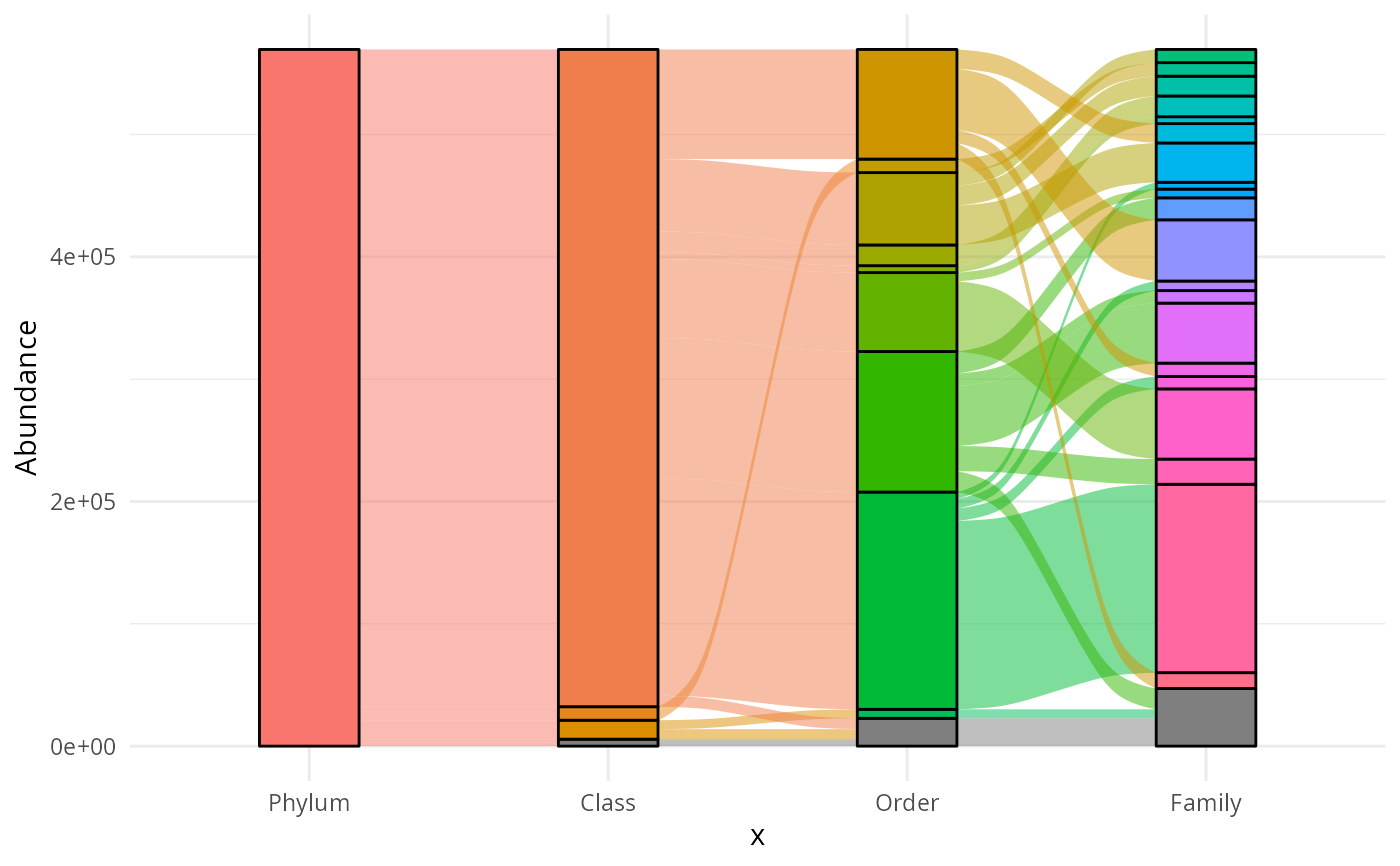 # \donttest{
if (requireNamespace("ggalluvial")) {
library(ggalluvial)
ggaluv_pq(data_fungi_mini)
ggaluv_pq(data_fungi_mini, type = "nb_taxa") +
geom_text(stat = "stratum", size = 1.8)
ggaluv_pq(data_fungi_mini,
wrap_factor = "Height",
by_sample = TRUE,
type = "nb_taxa"
) +
facet_wrap("Height")
ggaluv_pq(data_fungi_mini,
width = 0.9, min.size = 10,
type = "nb_taxa", taxa_ranks = c("Phylum", "Class", "Order", "Family", "Genus")
) + coord_flip() +
scale_x_discrete(limits = rev)
}
#> Warning: `group` has missing values; corresponding samples will be dropped
# \donttest{
if (requireNamespace("ggalluvial")) {
library(ggalluvial)
ggaluv_pq(data_fungi_mini)
ggaluv_pq(data_fungi_mini, type = "nb_taxa") +
geom_text(stat = "stratum", size = 1.8)
ggaluv_pq(data_fungi_mini,
wrap_factor = "Height",
by_sample = TRUE,
type = "nb_taxa"
) +
facet_wrap("Height")
ggaluv_pq(data_fungi_mini,
width = 0.9, min.size = 10,
type = "nb_taxa", taxa_ranks = c("Phylum", "Class", "Order", "Family", "Genus")
) + coord_flip() +
scale_x_discrete(limits = rev)
}
#> Warning: `group` has missing values; corresponding samples will be dropped
 # }
# }
