Venn diagram of phyloseq-class object using ggVennDiagram::ggVennDiagram function
Source: R/plot_functions.R
ggvenn_pq.RdNote that you can use ggplot2 function to customize the plot
for ex. + scale_fill_distiller(palette = "BuPu", direction = 1)
and + scale_x_continuous(expand = expansion(mult = 0.5)). See
examples.
Usage
ggvenn_pq(
physeq = NULL,
fact = NULL,
min_nb_seq = 0,
taxonomic_rank = NULL,
split_by = NULL,
add_nb_samples = TRUE,
add_nb_seq = FALSE,
rarefy_before_merging = FALSE,
rarefy_after_merging = FALSE,
rngseed = FALSE,
return_data_for_venn = FALSE,
verbose = TRUE,
type = "nb_taxa",
na_remove = TRUE,
...
)Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- fact
(required) Name of the factor to cluster samples by modalities. Need to be in
physeq@sam_data.- min_nb_seq
minimum number of sequences by OTUs by samples to take into count this OTUs in this sample. For example, if min_nb_seq=2,each value of 2 or less in the OTU table will not count in the venn diagram
- taxonomic_rank
Name (or number) of a taxonomic rank to count. If set to Null (the default) the number of OTUs is counted.
- split_by
Split into multiple plot using variable split_by. The name of a variable must be present in
sam_dataslot of the physeq object.- add_nb_samples
(logical, default TRUE) Add the number of samples to levels names
- add_nb_seq
(logical, default FALSE) Add the number of sequences to levels names
- rarefy_before_merging
Rarefy each sample before merging by the modalities of args
fact. Usephyloseq::rarefy_even_depth()function- rarefy_after_merging
Rarefy each sample after merging by the modalities of args
fact.- rngseed
(Optional). A single integer value passed to
phyloseq::rarefy_even_depth(), which is used to fix a seed for reproducibly random number generation (in this case, reproducibly random subsampling). If set to FALSE, then no fiddling with the RNG seed is performed, and it is up to the user to appropriately call set.seed beforehand to achieve reproducible results. Default is FALSE.- return_data_for_venn
(logical, default FALSE) If TRUE, the plot is not returned, but the resulting dataframe to plot with ggVennDiagram package is returned.
- verbose
(logical, default TRUE) If TRUE, prompt some messages.
- type
If "nb_taxa" (default), the number of taxa (ASV, OTU or taxonomic_rank if
taxonomic_rankis not NULL) is used in plot. If "nb_seq", the number of sequences is plotted.taxonomic_rankis never used if type = "nb_seq".- na_remove
(logical, default TRUE) If set to TRUE, remove samples with NA in the variables set in
factparam- ...
Other arguments for the
ggVennDiagram::ggVennDiagramfunction for ex.category.names.
Value
A ggplot2 plot representing Venn diagram of
modalities of the argument factor or if split_by is set a list
of plots.
Examples
if (requireNamespace("ggVennDiagram")) {
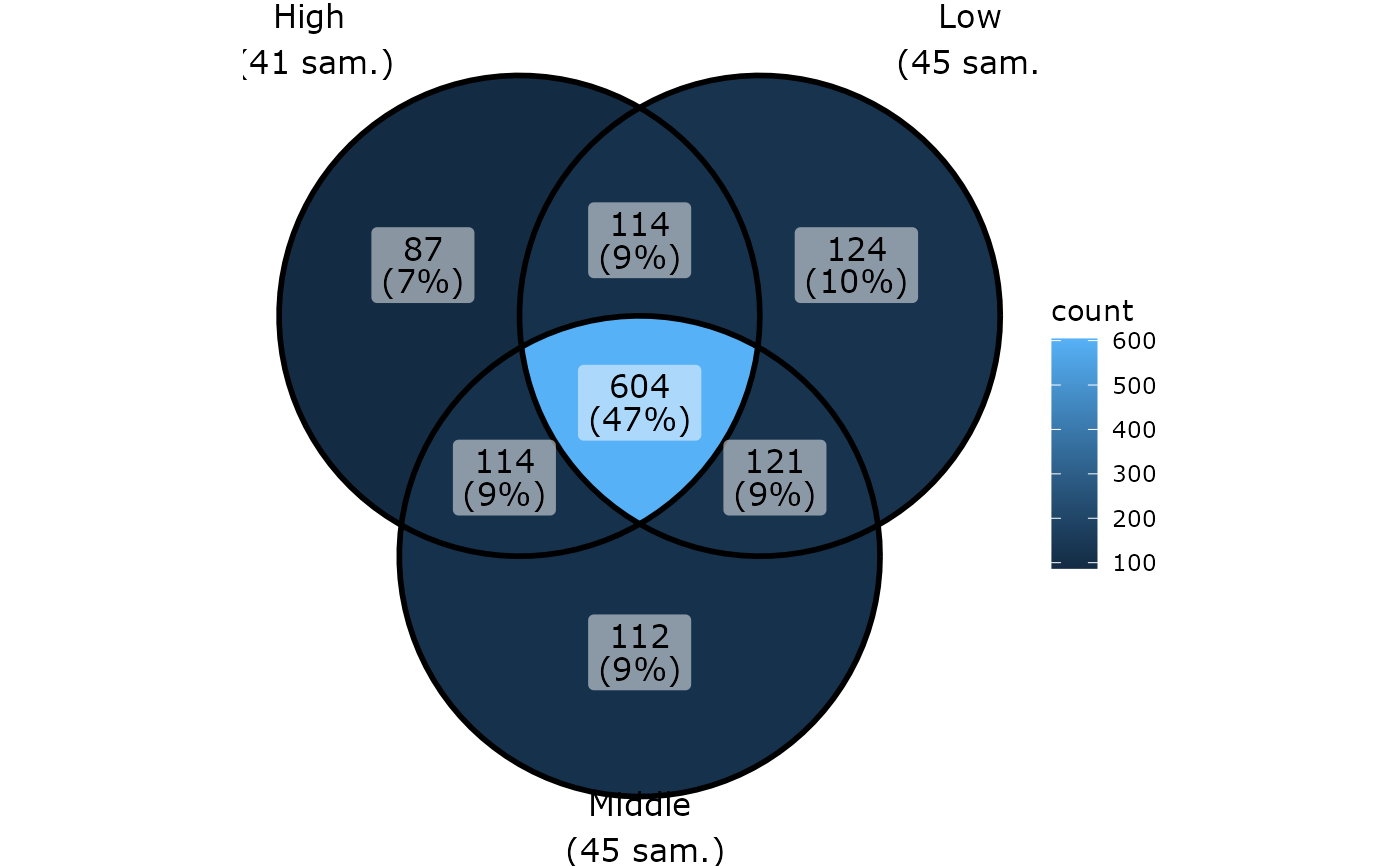
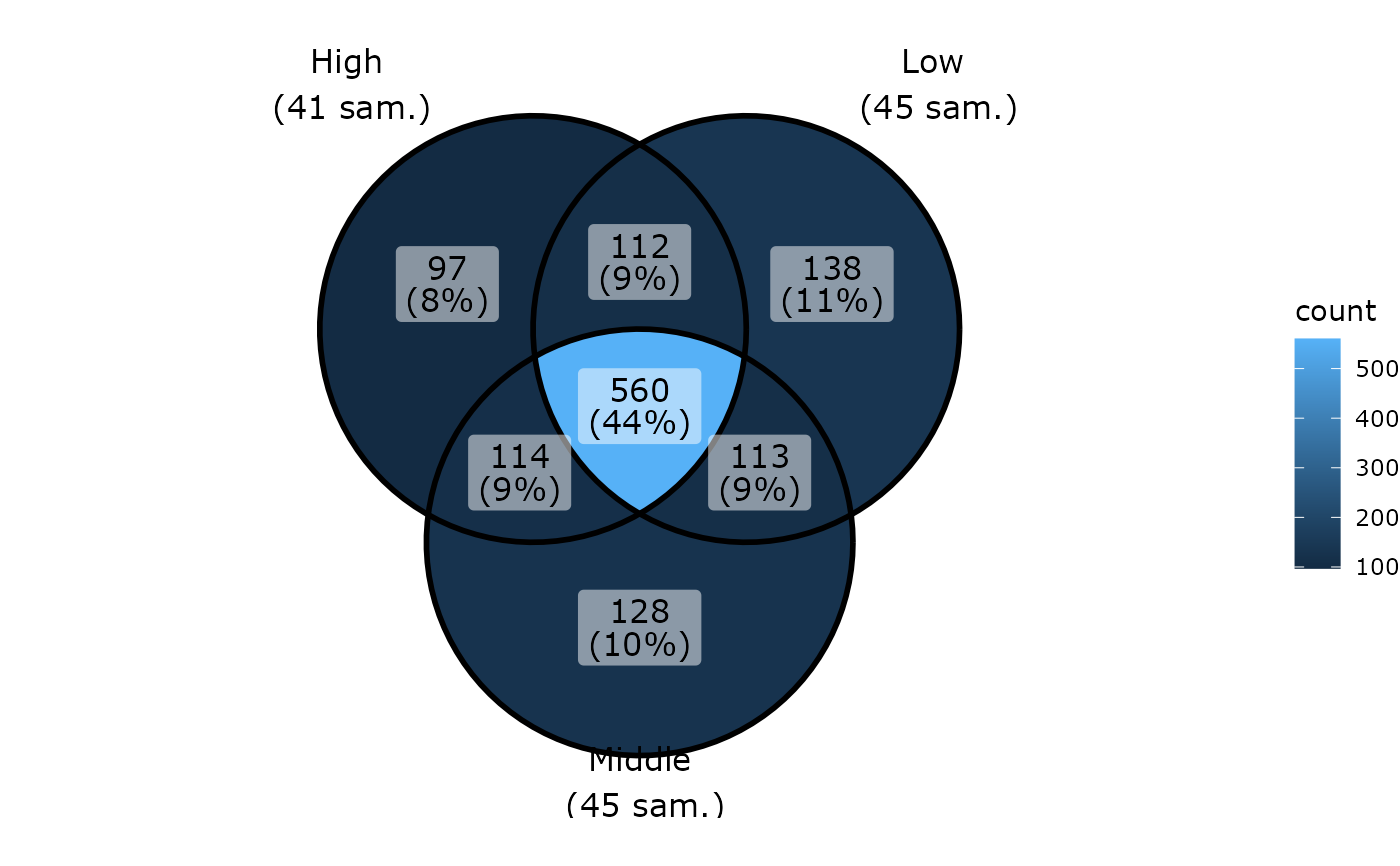
ggvenn_pq(data_fungi, fact = "Height")
}
#> 54 were discarded due to NA in variable fact
#> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 469 taxa and 0 samples.
 # \donttest{
if (requireNamespace("ggVennDiagram")) {
ggvenn_pq(data_fungi, fact = "Height") +
ggplot2::scale_fill_distiller(palette = "BuPu", direction = 1)
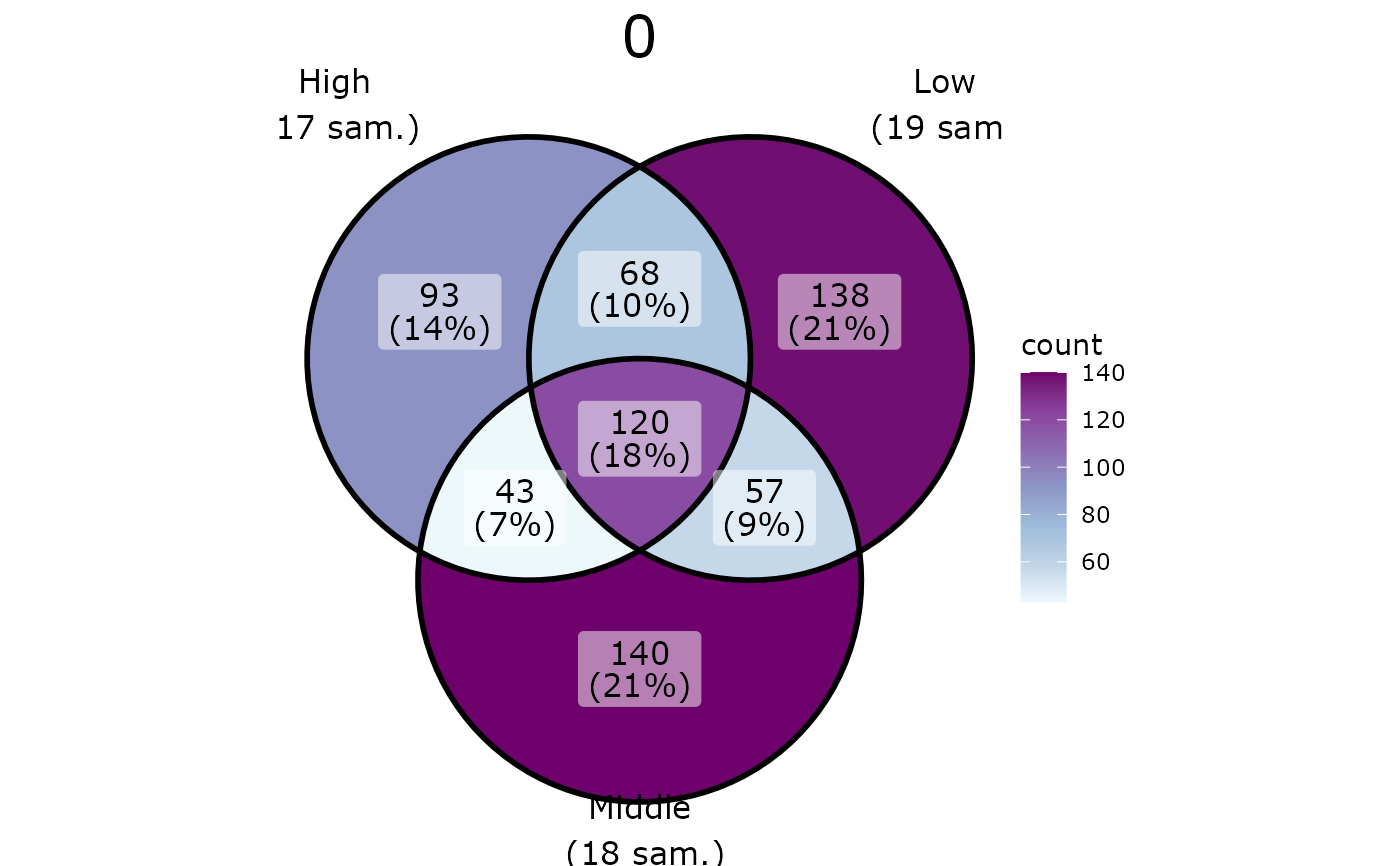
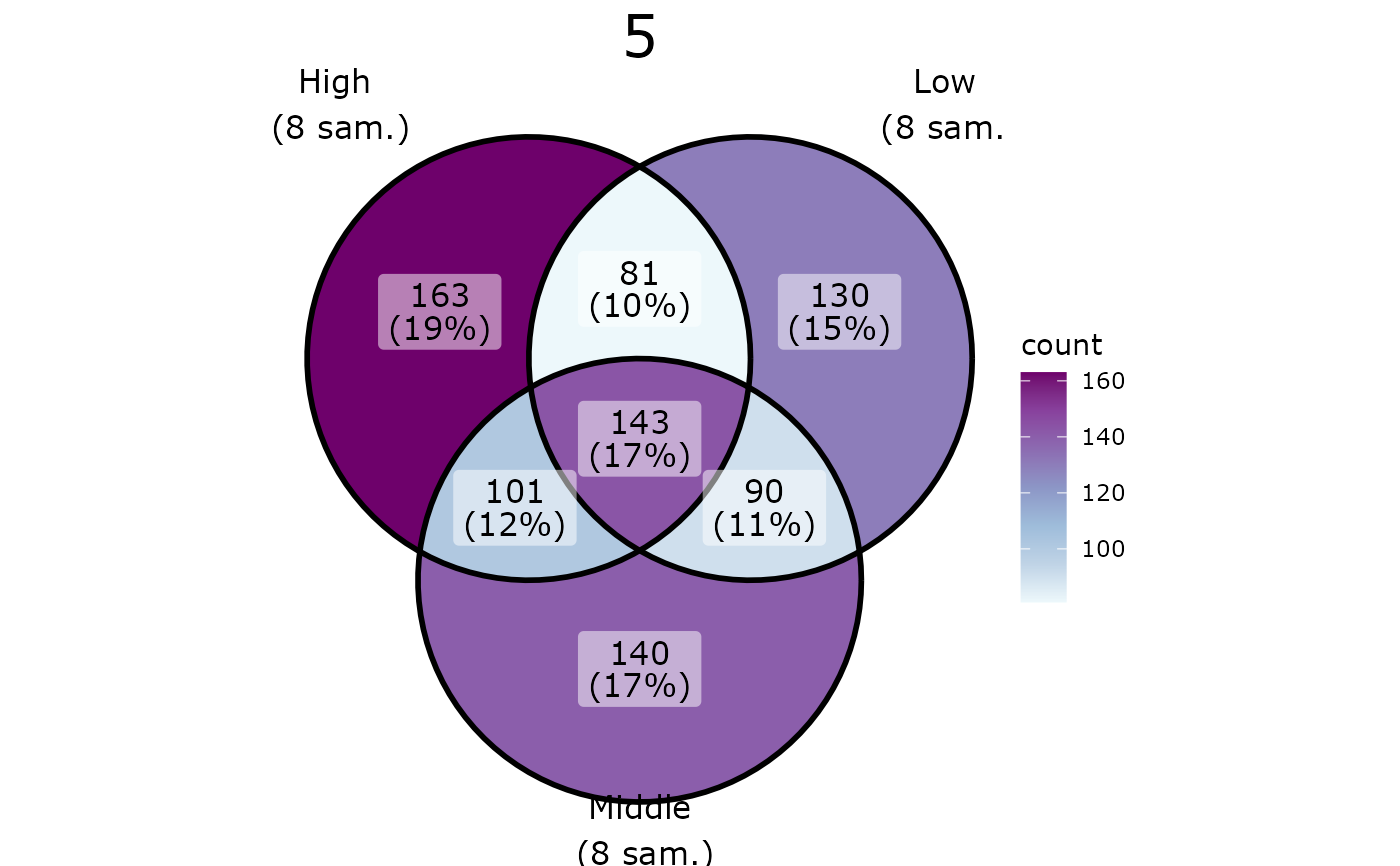
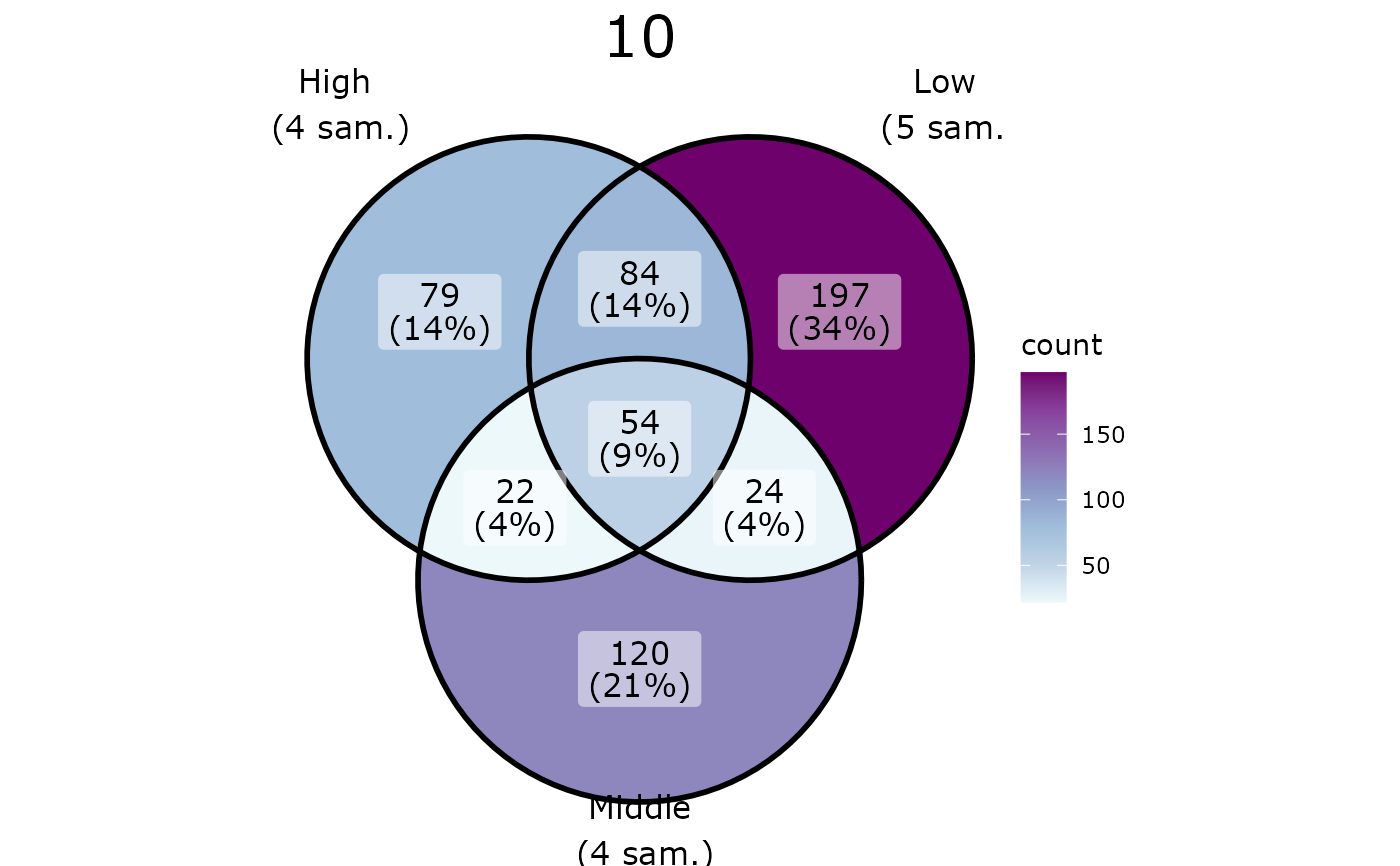
pl <- ggvenn_pq(data_fungi, fact = "Height", split_by = "Time")
for (i in seq_along(pl)) {
p <- pl[[i]] +
scale_fill_distiller(palette = "BuPu", direction = 1) +
theme(plot.title = element_text(hjust = 0.5, size = 22))
print(p)
}
data_fungi2 <- subset_samples(data_fungi, data_fungi@sam_data$Tree_name == "A10-005" |
data_fungi@sam_data$Height %in% c("Low", "High"))
ggvenn_pq(data_fungi2, fact = "Height")
ggvenn_pq(data_fungi2, fact = "Height", type = "nb_seq")
ggvenn_pq(data_fungi, fact = "Height", add_nb_seq = TRUE, set_size = 4)
ggvenn_pq(data_fungi, fact = "Height", rarefy_before_merging = TRUE)
ggvenn_pq(data_fungi, fact = "Height", rarefy_after_merging = TRUE) +
scale_x_continuous(expand = expansion(mult = 0.5))
# For more flexibility, you can save the dataset for more precise construction
# with ggplot2 and ggVennDiagramm
# (https://gaospecial.github.io/ggVennDiagram/articles/fully-customed.html)
res_venn <- ggvenn_pq(data_fungi, fact = "Height", return_data_for_venn = TRUE)
ggplot() +
# 1. region count layer
geom_polygon(aes(X, Y, group = id, fill = name),
data = ggVennDiagram::venn_regionedge(res_venn)
) +
scale_fill_manual(values = funky_color(7)) +
# 2. set edge layer
geom_path(aes(X, Y, color = id, group = id),
data = ggVennDiagram::venn_setedge(res_venn),
show.legend = FALSE, linewidth = 2
) +
scale_color_manual(values = c("red", "red", "blue")) +
# 3. set label layer
geom_text(aes(X, Y, label = name),
data = ggVennDiagram::venn_setlabel(res_venn)
) +
# 4. region label layer
geom_label(
aes(X, Y, label = paste0(
count, " (",
scales::percent(count / sum(count), accuracy = 2), ")"
)),
data = ggVennDiagram::venn_regionlabel(res_venn)
) +
theme_void()
}
#> 54 were discarded due to NA in variable fact
#> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 469 taxa and 0 samples.
#> 54 were discarded due to NA in variable fact
#> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 469 taxa and 0 samples.
#> Cleaning suppress 335 taxa and 0 samples.
#> Cleaning suppress 276 taxa and 0 samples.
#> Cleaning suppress 299 taxa and 0 samples.
#> Cleaning suppress 360 taxa and 0 samples.
#> Cleaning suppress 404 taxa and 0 samples.
#> Cleaning suppress 374 taxa and 0 samples.
#> Cleaning suppress 341 taxa and 0 samples.
#> Cleaning suppress 221 taxa and 0 samples.
#> Cleaning suppress 360 taxa and 0 samples.
#> Cleaning suppress 343 taxa and 0 samples.
#> Cleaning suppress 218 taxa and 0 samples.
#> Cleaning suppress 408 taxa and 0 samples.
#> Two modalities differ greatly (more than x2) in their number of sequences (159635 vs 53355). You may be interested by the parameter rarefy_after_merging
# \donttest{
if (requireNamespace("ggVennDiagram")) {
ggvenn_pq(data_fungi, fact = "Height") +
ggplot2::scale_fill_distiller(palette = "BuPu", direction = 1)
pl <- ggvenn_pq(data_fungi, fact = "Height", split_by = "Time")
for (i in seq_along(pl)) {
p <- pl[[i]] +
scale_fill_distiller(palette = "BuPu", direction = 1) +
theme(plot.title = element_text(hjust = 0.5, size = 22))
print(p)
}
data_fungi2 <- subset_samples(data_fungi, data_fungi@sam_data$Tree_name == "A10-005" |
data_fungi@sam_data$Height %in% c("Low", "High"))
ggvenn_pq(data_fungi2, fact = "Height")
ggvenn_pq(data_fungi2, fact = "Height", type = "nb_seq")
ggvenn_pq(data_fungi, fact = "Height", add_nb_seq = TRUE, set_size = 4)
ggvenn_pq(data_fungi, fact = "Height", rarefy_before_merging = TRUE)
ggvenn_pq(data_fungi, fact = "Height", rarefy_after_merging = TRUE) +
scale_x_continuous(expand = expansion(mult = 0.5))
# For more flexibility, you can save the dataset for more precise construction
# with ggplot2 and ggVennDiagramm
# (https://gaospecial.github.io/ggVennDiagram/articles/fully-customed.html)
res_venn <- ggvenn_pq(data_fungi, fact = "Height", return_data_for_venn = TRUE)
ggplot() +
# 1. region count layer
geom_polygon(aes(X, Y, group = id, fill = name),
data = ggVennDiagram::venn_regionedge(res_venn)
) +
scale_fill_manual(values = funky_color(7)) +
# 2. set edge layer
geom_path(aes(X, Y, color = id, group = id),
data = ggVennDiagram::venn_setedge(res_venn),
show.legend = FALSE, linewidth = 2
) +
scale_color_manual(values = c("red", "red", "blue")) +
# 3. set label layer
geom_text(aes(X, Y, label = name),
data = ggVennDiagram::venn_setlabel(res_venn)
) +
# 4. region label layer
geom_label(
aes(X, Y, label = paste0(
count, " (",
scales::percent(count / sum(count), accuracy = 2), ")"
)),
data = ggVennDiagram::venn_regionlabel(res_venn)
) +
theme_void()
}
#> 54 were discarded due to NA in variable fact
#> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 469 taxa and 0 samples.
#> 54 were discarded due to NA in variable fact
#> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 469 taxa and 0 samples.
#> Cleaning suppress 335 taxa and 0 samples.
#> Cleaning suppress 276 taxa and 0 samples.
#> Cleaning suppress 299 taxa and 0 samples.
#> Cleaning suppress 360 taxa and 0 samples.
#> Cleaning suppress 404 taxa and 0 samples.
#> Cleaning suppress 374 taxa and 0 samples.
#> Cleaning suppress 341 taxa and 0 samples.
#> Cleaning suppress 221 taxa and 0 samples.
#> Cleaning suppress 360 taxa and 0 samples.
#> Cleaning suppress 343 taxa and 0 samples.
#> Cleaning suppress 218 taxa and 0 samples.
#> Cleaning suppress 408 taxa and 0 samples.
#> Two modalities differ greatly (more than x2) in their number of sequences (159635 vs 53355). You may be interested by the parameter rarefy_after_merging
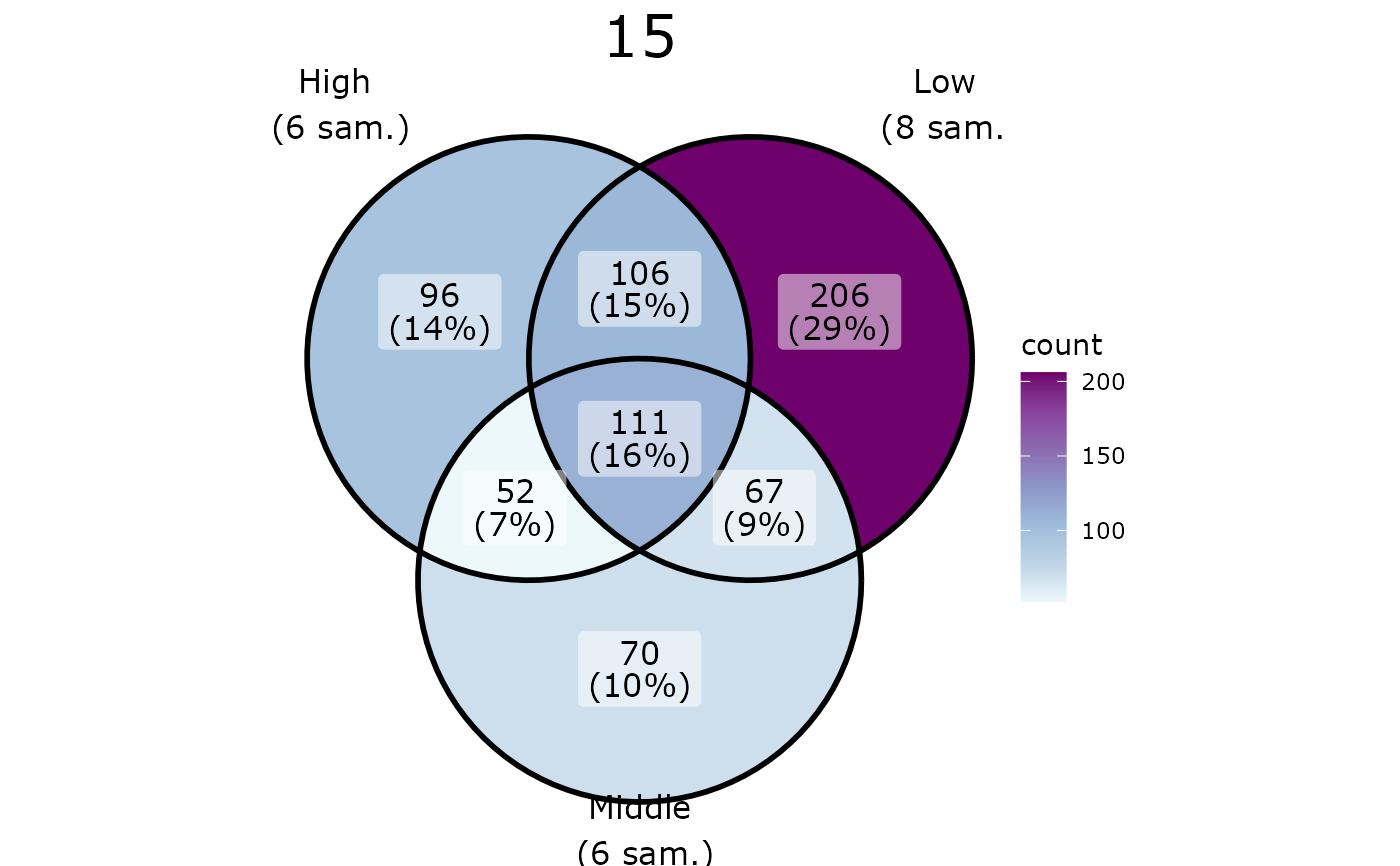 #> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 1366 taxa and 0 samples.
#> Two modalities differ greatly (more than x2) in their number of sequences (432919 vs 3961). You may be interested by the parameter rarefy_after_merging
#> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 1366 taxa and 0 samples.
#> Two modalities differ greatly (more than x2) in their number of sequences (432919 vs 3961). You may be interested by the parameter rarefy_after_merging
#> 54 were discarded due to NA in variable fact
#> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 469 taxa and 0 samples.
#> 54 were discarded due to NA in variable fact
#> You set `rngseed` to FALSE. Make sure you've set & recorded
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> ...
#> You set `rngseed` to FALSE. Make sure you've set & recorded
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> ...
#> 1141OTUs were removed because they are no longer
#> present in any sample after random subsampling
#> ...
#> Cleaning suppress 171 taxa and 0 samples.
#> Cleaning suppress 146 taxa and 0 samples.
#> Cleaning suppress 153 taxa and 0 samples.
#> 54 were discarded due to NA in variable fact
#> You set `rngseed` to FALSE. Make sure you've set & recorded
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> ...
#> You set `rngseed` to FALSE. Make sure you've set & recorded
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> ...
#> 157OTUs were removed because they are no longer
#> present in any sample after random subsampling
#> ...
#> Cleaning suppress 385 taxa and 0 samples.
#> Cleaning suppress 342 taxa and 0 samples.
#> Cleaning suppress 340 taxa and 0 samples.
#> 54 were discarded due to NA in variable fact
#> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 469 taxa and 0 samples.
#> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 1366 taxa and 0 samples.
#> Two modalities differ greatly (more than x2) in their number of sequences (432919 vs 3961). You may be interested by the parameter rarefy_after_merging
#> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 1366 taxa and 0 samples.
#> Two modalities differ greatly (more than x2) in their number of sequences (432919 vs 3961). You may be interested by the parameter rarefy_after_merging
#> 54 were discarded due to NA in variable fact
#> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 469 taxa and 0 samples.
#> 54 were discarded due to NA in variable fact
#> You set `rngseed` to FALSE. Make sure you've set & recorded
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> ...
#> You set `rngseed` to FALSE. Make sure you've set & recorded
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> ...
#> 1141OTUs were removed because they are no longer
#> present in any sample after random subsampling
#> ...
#> Cleaning suppress 171 taxa and 0 samples.
#> Cleaning suppress 146 taxa and 0 samples.
#> Cleaning suppress 153 taxa and 0 samples.
#> 54 were discarded due to NA in variable fact
#> You set `rngseed` to FALSE. Make sure you've set & recorded
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> ...
#> You set `rngseed` to FALSE. Make sure you've set & recorded
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> ...
#> 157OTUs were removed because they are no longer
#> present in any sample after random subsampling
#> ...
#> Cleaning suppress 385 taxa and 0 samples.
#> Cleaning suppress 342 taxa and 0 samples.
#> Cleaning suppress 340 taxa and 0 samples.
#> 54 were discarded due to NA in variable fact
#> Cleaning suppress 501 taxa and 0 samples.
#> Cleaning suppress 457 taxa and 0 samples.
#> Cleaning suppress 469 taxa and 0 samples.
 # }
# }
