Performs graph-based permutation tests on phyloseq object
Source:R/beta_div_test.R
graph_test_pq.RdA wrapper of phyloseqGraphTest::graph_perm_test() for quick plot with
important statistics
Usage
graph_test_pq(
physeq,
fact,
merge_sample_by = NULL,
nperm = 999,
return_plot = TRUE,
title = "Graph Test",
na_remove = FALSE,
...
)Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- fact
(required) Name of the factor to cluster samples by modalities. Need to be in
physeq@sam_data. This should be a factor with two or more levels.- merge_sample_by
a vector to determine which samples to merge using
merge_samples2()function. Need to be inphyseq@sam_data- nperm
(int) The number of permutations to perform.
- return_plot
(logical) Do we return only the result of the test, or do we plot the result?
- title
The title of the Graph.
- na_remove
(logical, default FALSE) If set to TRUE, remove samples with NA in the variables set in formula.
- ...
Other params for be passed on to
phyloseqGraphTest::graph_perm_test()function
Value
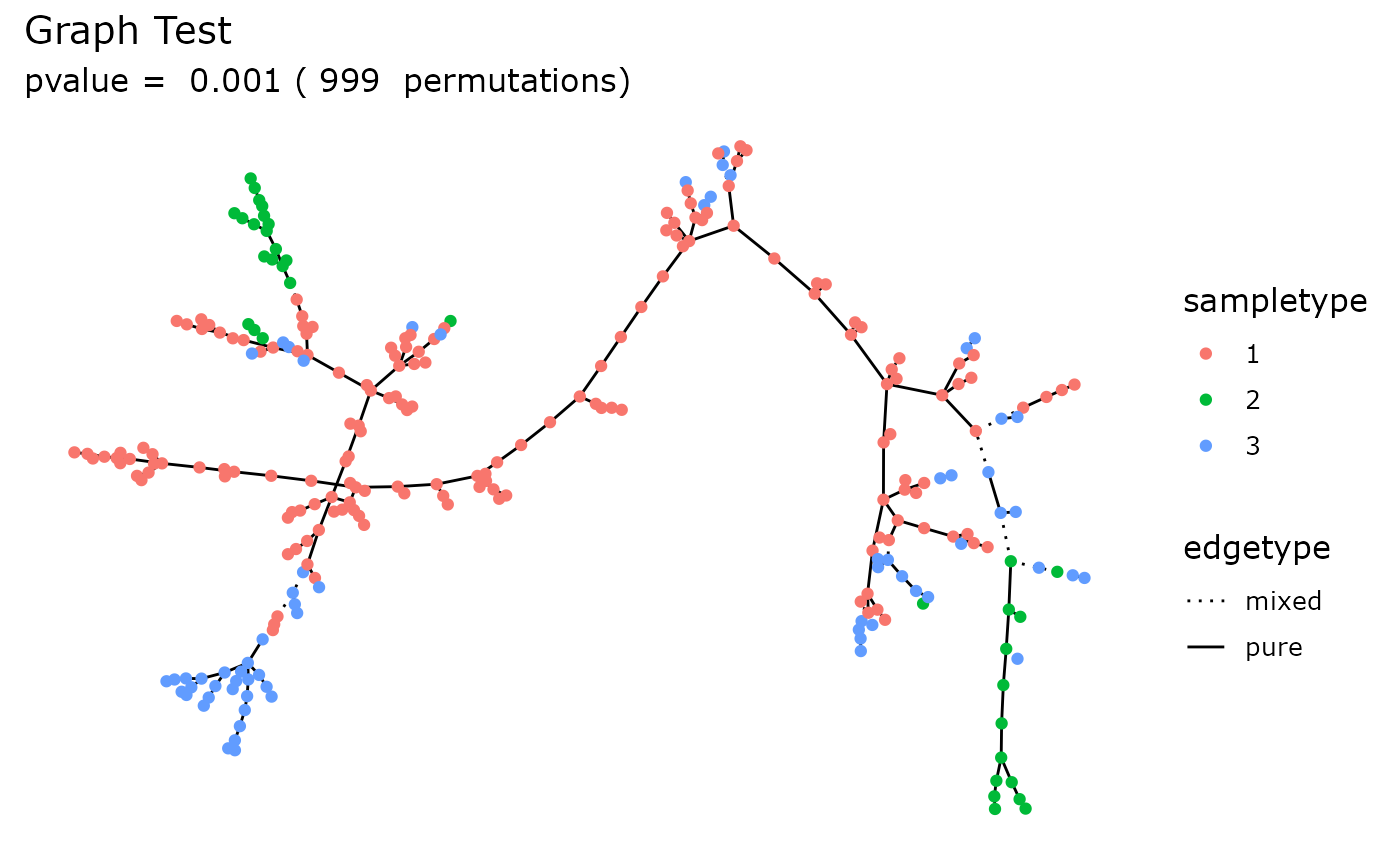
A ggplot2 plot with a subtitle indicating the pvalue
and the number of permutations
Details
This function is mainly a wrapper of the work of others.
Please cite phyloseqGraphTest package.
Examples
# \donttest{
if (requireNamespace("phyloseqGraphTest")) {
data(enterotype)
graph_test_pq(enterotype, fact = "SeqTech")
graph_test_pq(enterotype, fact = "Enterotype", na_remove = TRUE)
}
#> 9 were discarded due to NA in variables present in formula.
 # }
# }
