A wrapper for the indicspecies::multipatt() function in the case of
physeq object.
Usage
multipatt_pq(
physeq,
fact,
p_adjust_method = "BH",
pval = 0.05,
control = permute::how(nperm = 999),
...
)Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- fact
(required) Name of the factor in
physeq@sam_dataused to plot different lines- p_adjust_method
(chr, default "BH"): the method used to adjust p-value
- pval
(int, default 0.05): the value to determine the significance of LCBD
- control
see
?indicspecies::multipatt()- ...
Additional arguments passed on to
indicspecies::multipatt()function
Details
This function is mainly a wrapper of the work of others.
Please make a reference to indicspecies::multipatt() if you
use this function.
Examples
if (requireNamespace("indicspecies")) {
data(data_fungi)
data_fungi_ab <- subset_taxa_pq(data_fungi, taxa_sums(data_fungi) > 10000)
multipatt_pq(subset_samples(data_fungi_ab, !is.na(Time)), fact = "Time")
}
#> Cleaning suppress 0 taxa ( ) and 15 sample(s) ( BE9-006-B_S27_MERGED.fastq.gz / C21-NV1-M_S64_MERGED.fastq.gz / DJ2-008-B_S87_MERGED.fastq.gz / DY5-004-H_S97_MERGED.fastq.gz / DY5-004-M_S98_MERGED.fastq.gz / E9-009-B_S100_MERGED.fastq.gz / E9-009-H_S101_MERGED.fastq.gz / N22-001-B_S129_MERGED.fastq.gz / O20-X-B_S139_MERGED.fastq.gz / O21-007-M_S144_MERGED.fastq.gz / R28-008-H_S159_MERGED.fastq.gz / R28-008-M_S160_MERGED.fastq.gz / W26-001-M_S167_MERGED.fastq.gz / Y29-007-H_S182_MERGED.fastq.gz / Y29-007-M_S183_MERGED.fastq.gz ).
#> Number of non-matching ASV 0
#> Number of matching ASV 1420
#> Number of filtered-out ASV 1385
#> Number of kept ASV 35
#> Number of kept samples 170
#> Taxa are now in columns.
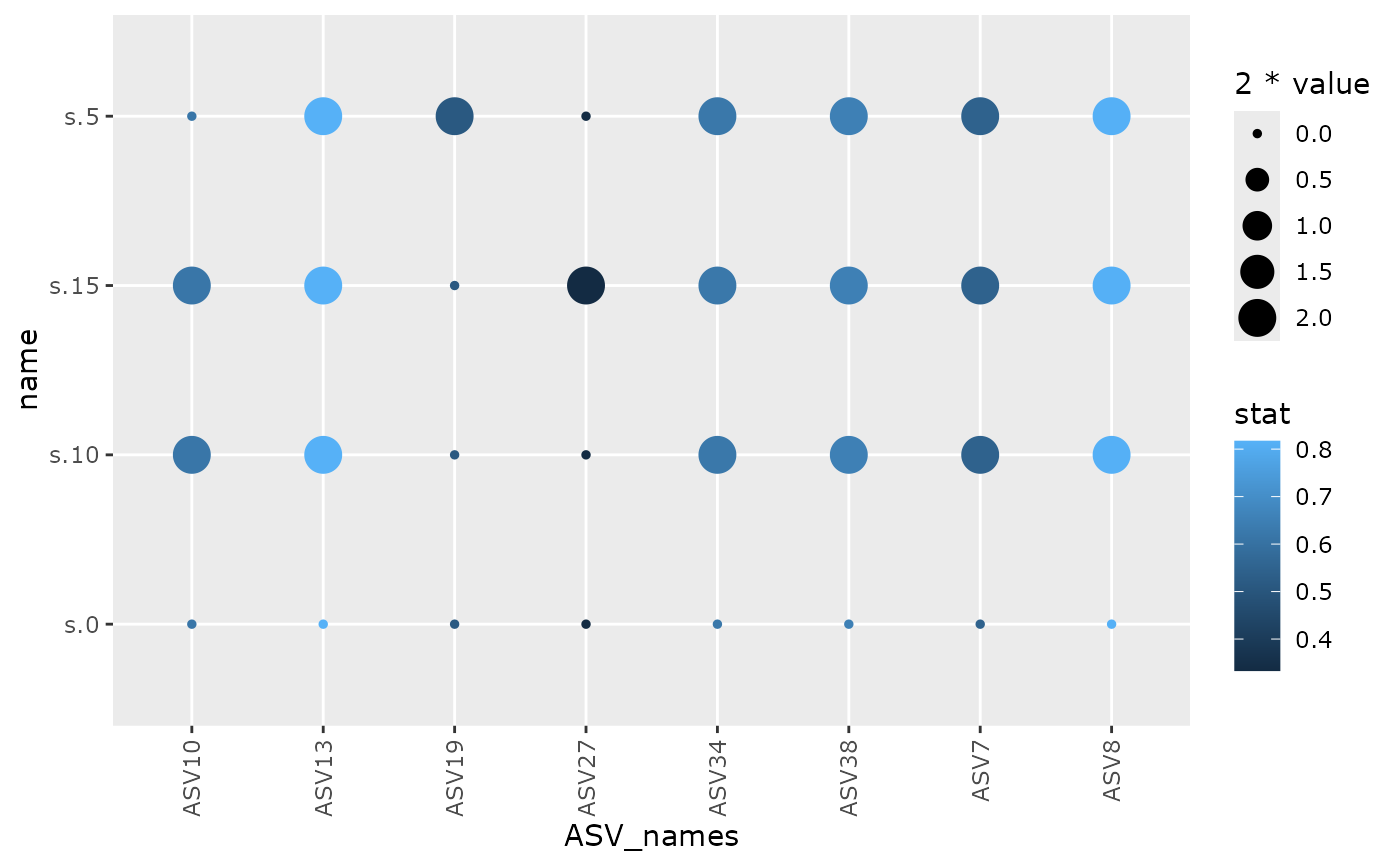 # \donttest{
if (requireNamespace("indicspecies")) {
multipatt_pq(subset_samples(data_fungi_ab, !is.na(Time)),
fact = "Time",
max.order = 1, control = permute::how(nperm = 99)
)
}
#> Taxa are now in columns.
# \donttest{
if (requireNamespace("indicspecies")) {
multipatt_pq(subset_samples(data_fungi_ab, !is.na(Time)),
fact = "Time",
max.order = 1, control = permute::how(nperm = 99)
)
}
#> Taxa are now in columns.
 # }
# }
