Plot and test local contributions to beta diversity (LCBD) of samples
Source:R/beta_div_test.R
plot_LCBD_pq.RdA wrapper for the adespatial::beta.div() function in the case of physeq
object.
Usage
plot_LCBD_pq(
physeq,
p_adjust_method = "BH",
pval = 0.05,
sam_variables = NULL,
only_plot_significant = TRUE,
...
)Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- p_adjust_method
(chr, default "BH"): the method used to adjust p-value
- pval
(int, default 0.05): the value to determine the significance of LCBD
- sam_variables
A vector of variable names present in the
sam_dataslot to plot alongside the LCBD value- only_plot_significant
(logical, default TRUE) Do we plot all LCBD values or only the significant ones
- ...
Additional arguments passed on to
adespatial::beta.div()function
Details
This function is mainly a wrapper of the work of others.
Please make a reference to vegan::beta.div() if you
use this function.
Examples
data(data_fungi)
if (requireNamespace("adespatial")) {
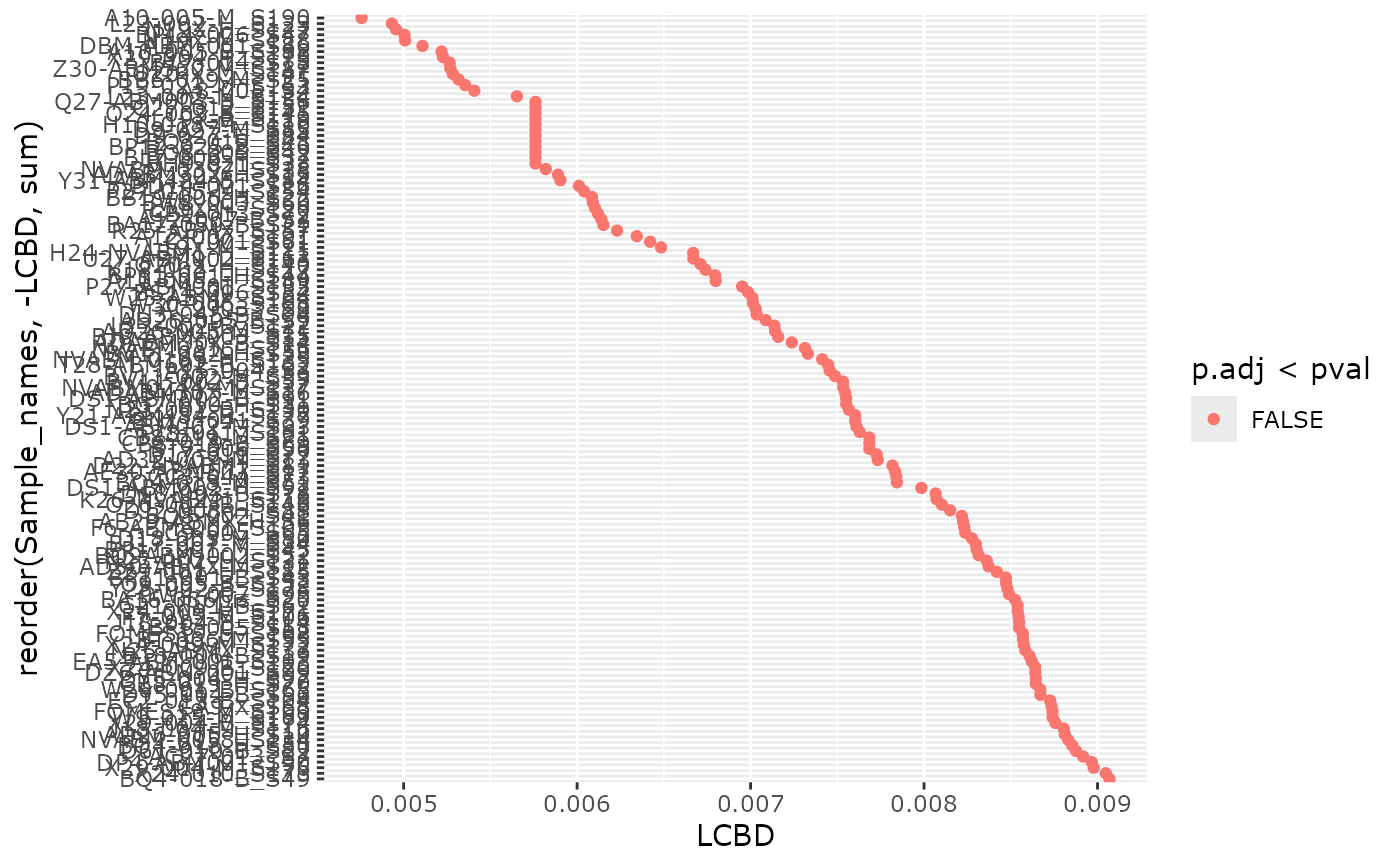
plot_LCBD_pq(data_fungi_mini,
nperm = 100, only_plot_significant = FALSE,
pval = 0.2
)
}
#> Taxa are now in columns.
 # \donttest{
if (requireNamespace("adespatial")) {
plot_LCBD_pq(data_fungi_mini,
nperm = 100, only_plot_significant = TRUE,
pval = 0.2
)
if (requireNamespace("patchwork")) {
plot_LCBD_pq(data_fungi_mini,
nperm = 100, only_plot_significant = FALSE,
sam_variables = c("Time", "Height")
)
plot_LCBD_pq(data_fungi_mini,
nperm = 100, only_plot_significant = TRUE, pval = 0.2,
sam_variables = c("Time", "Height", "Tree_name")
) &
theme(
legend.key.size = unit(0.4, "cm"),
legend.text = element_text(size = 10),
axis.title.x = element_text(size = 6)
)
}
}
#> Taxa are now in columns.
#> Taxa are now in columns.
#> Taxa are now in columns.
# \donttest{
if (requireNamespace("adespatial")) {
plot_LCBD_pq(data_fungi_mini,
nperm = 100, only_plot_significant = TRUE,
pval = 0.2
)
if (requireNamespace("patchwork")) {
plot_LCBD_pq(data_fungi_mini,
nperm = 100, only_plot_significant = FALSE,
sam_variables = c("Time", "Height")
)
plot_LCBD_pq(data_fungi_mini,
nperm = 100, only_plot_significant = TRUE, pval = 0.2,
sam_variables = c("Time", "Height", "Tree_name")
) &
theme(
legend.key.size = unit(0.4, "cm"),
legend.text = element_text(size = 10),
axis.title.x = element_text(size = 6)
)
}
}
#> Taxa are now in columns.
#> Taxa are now in columns.
#> Taxa are now in columns.
 # }
# }
