Usage
plot_edgeR_pq(
physeq,
contrast = NULL,
pval = 0.05,
taxolev = "Genus",
color_tax = "Phylum",
verbose = TRUE,
...
)Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- contrast
(required) This argument specifies what comparison to extract from the object to build a results table. See
resultsman page for more details.- pval
(default: 0.05) the significance cutoff used for optimizing the independent filtering. If the adjusted p-value cutoff (FDR) will be a value other than 0.05, pval should be set to that value.
- taxolev
taxonomic level of interest
- color_tax
taxonomic level used for color assignation
- verbose
(logical): whether the function print some information during the computation
- ...
Value
A ggplot2 plot representing edgeR results
Examples
data("GlobalPatterns", package = "phyloseq")
GP_archae <- subset_taxa(GlobalPatterns, GlobalPatterns@tax_table[, 1] == "Archaea")
#> Found more than one class "phylo" in cache; using the first, from namespace 'phyloseq'
#> Also defined by ‘RNeXML’
#> Found more than one class "phylo" in cache; using the first, from namespace 'phyloseq'
#> Also defined by ‘RNeXML’
#> Found more than one class "phylo" in cache; using the first, from namespace 'phyloseq'
#> Also defined by ‘RNeXML’
#> Found more than one class "phylo" in cache; using the first, from namespace 'phyloseq'
#> Also defined by ‘RNeXML’
# \donttest{
if (requireNamespace("edgeR")) {
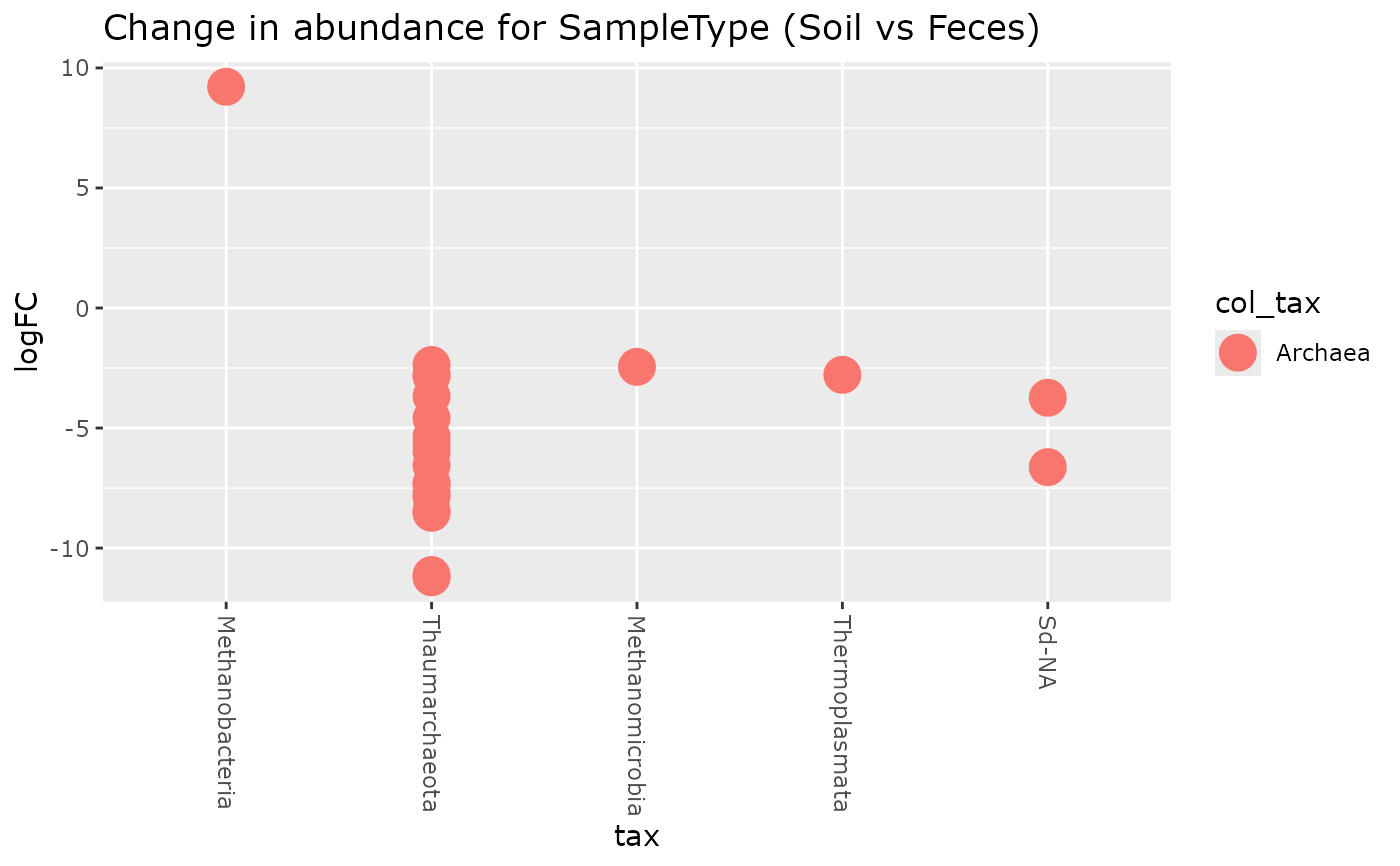
plot_edgeR_pq(GP_archae, c("SampleType", "Soil", "Feces"),
color_tax = "Kingdom"
)
plot_edgeR_pq(GP_archae, c("SampleType", "Soil", "Feces"),
taxolev = "Class", color_tax = "Kingdom"
)
}
#> Conversion to edgeR format
#> Found more than one class "phylo" in cache; using the first, from namespace 'phyloseq'
#> Also defined by ‘RNeXML’
#> Perform edgeR binary test
#> Conversion to edgeR format
#> Found more than one class "phylo" in cache; using the first, from namespace 'phyloseq'
#> Also defined by ‘RNeXML’
#> Perform edgeR binary test
 # }
# }
