Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- lvl1
(required) Name of the first (higher) taxonomic rank of interest
- lvl2
(required) Name of the second (lower) taxonomic rank of interest
- nb_seq
(logical; default TRUE) If set to FALSE, only the number of ASV is count. Concretely, physeq otu_table is transformed in a binary otu_table (each value different from zero is set to one)
- log10trans
(logical, default TRUE) If TRUE, the number of sequences (or ASV if nb_seq = FALSE) is log10 transformed.
- plot_legend
(logical, default FALSE) If TRUE, plot che legend of color for lvl 1
- ...
Additional arguments passed on to
treemapify::geom_treemap()function.
Examples
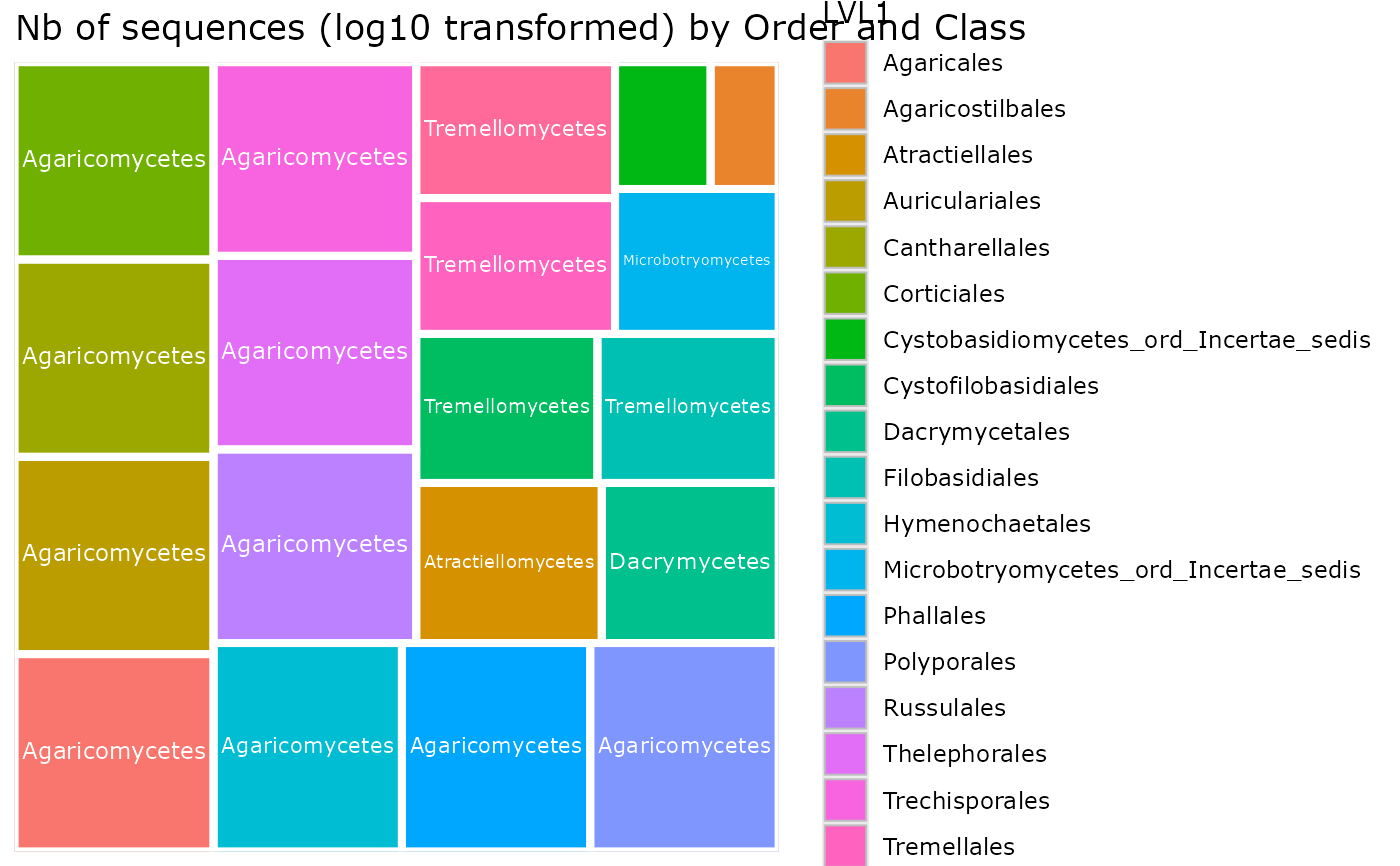
data(data_fungi_sp_known)
if (requireNamespace("treemapify")) {
treemap_pq(
clean_pq(subset_taxa(
data_fungi_sp_known,
Phylum == "Basidiomycota"
)),
"Order", "Class",
plot_legend = TRUE
)
}
#> Loading required namespace: treemapify
#> Cleaning suppress 0 taxa and 5 samples.
 # \donttest{
if (requireNamespace("treemapify")) {
treemap_pq(
clean_pq(subset_taxa(
data_fungi_sp_known,
Phylum == "Basidiomycota"
)),
"Order", "Class",
log10trans = FALSE
)
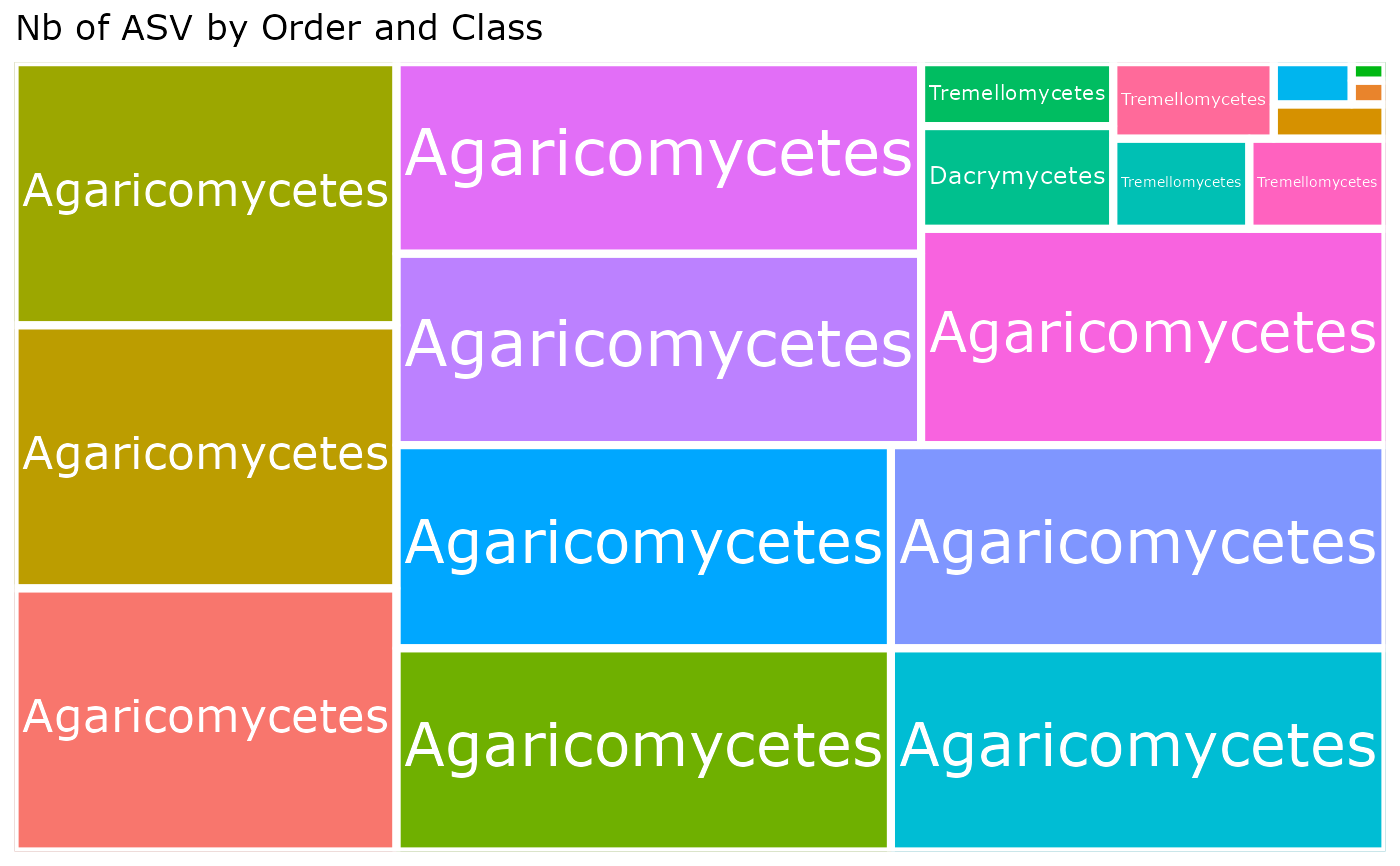
treemap_pq(
clean_pq(subset_taxa(
data_fungi_sp_known,
Phylum == "Basidiomycota"
)),
"Order", "Class",
nb_seq = FALSE, log10trans = FALSE
)
}
#> Cleaning suppress 0 taxa and 5 samples.
#> Cleaning suppress 0 taxa and 5 samples.
# \donttest{
if (requireNamespace("treemapify")) {
treemap_pq(
clean_pq(subset_taxa(
data_fungi_sp_known,
Phylum == "Basidiomycota"
)),
"Order", "Class",
log10trans = FALSE
)
treemap_pq(
clean_pq(subset_taxa(
data_fungi_sp_known,
Phylum == "Basidiomycota"
)),
"Order", "Class",
nb_seq = FALSE, log10trans = FALSE
)
}
#> Cleaning suppress 0 taxa and 5 samples.
#> Cleaning suppress 0 taxa and 5 samples.
 # }
# }
