Computes a manifold approximation and projection (UMAP) for phyloseq object
Source:R/plot_functions.R
umap_pq.RdArguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- pkg
Which R packages to use, either "umap" or "uwot".
- ...
Additional arguments passed on to
umap::umap()oruwot::umap2()function. For examplen_neighborsset the number of nearest neighbors (Default 15). Seeumap::umap.defaults()oruwot::umap2()for the list of parameters and default values.
Details
This function is mainly a wrapper of the work of others.
Please make a reference to umap::umap() if you
use this function.
Examples
library("umap")
df_umap <- umap_pq(data_fungi_mini)
#> Taxa are now in columns.
#> Taxa are now in rows.
#> Joining with `by = join_by(Sample)`
#> Warning: The `x` argument of `as_tibble.matrix()` must have unique column names if
#> `.name_repair` is omitted as of tibble 2.0.0.
#> ℹ Using compatibility `.name_repair`.
#> ℹ The deprecated feature was likely used in the MiscMetabar package.
#> Please report the issue at
#> <https://github.com/adrientaudiere/MiscMetabar/issues>.
#> Joining with `by = join_by(Sample)`
ggplot(df_umap, aes(x = x_umap, y = y_umap, col = Height)) +
geom_point(size = 2)
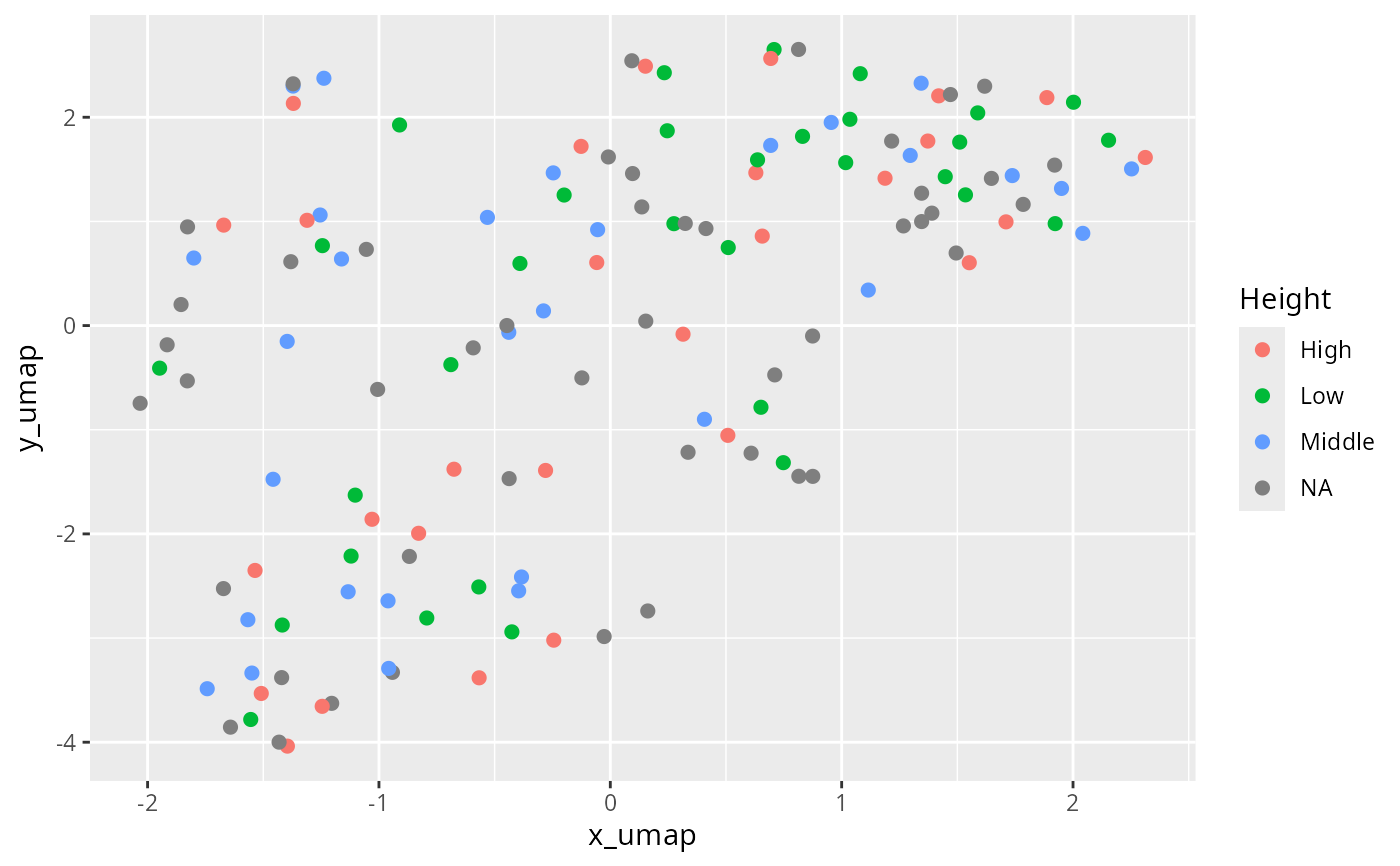 # \donttest{
library(patchwork)
physeq <- data_fungi_mini
res_tsne <- tsne_pq(data_fungi_mini)
df_umap_tsne <- df_umap
df_umap_tsne$x_tsne <- res_tsne$Y[, 1]
df_umap_tsne$y_tsne <- res_tsne$Y[, 2]
((ggplot(df_umap, aes(x = x_umap, y = y_umap, col = Height)) +
geom_point(size = 2) +
ggtitle("UMAP")) + (plot_ordination(physeq,
ordination =
ordinate(physeq, method = "PCoA", distance = "bray"), color = "Height"
)) +
ggtitle("PCoA")) /
((ggplot(df_umap_tsne, aes(x = x_tsne, y = y_tsne, col = Height)) +
geom_point(size = 2) +
ggtitle("tsne")) +
(plot_ordination(physeq,
ordination = ordinate(physeq, method = "NMDS", distance = "bray"),
color = "Height"
) +
ggtitle("NMDS"))) +
patchwork::plot_layout(guides = "collect")
#> Square root transformation
#> Wisconsin double standardization
#> Run 0 stress 0.1842444
#> Run 1 stress 0.1864672
#> Run 2 stress 0.1816642
#> ... New best solution
#> ... Procrustes: rmse 0.06035663 max resid 0.2763356
#> Run 3 stress 0.1805408
#> ... New best solution
#> ... Procrustes: rmse 0.0696832 max resid 0.2635835
#> Run 4 stress 0.1918613
#> Run 5 stress 0.1837193
#> Run 6 stress 0.1856894
#> Run 7 stress 0.1854912
#> Run 8 stress 0.1855901
#> Run 9 stress 0.1828179
#> Run 10 stress 0.1797561
#> ... New best solution
#> ... Procrustes: rmse 0.05144056 max resid 0.2994319
#> Run 11 stress 0.1861731
#> Run 12 stress 0.1861394
#> Run 13 stress 0.1881243
#> Run 14 stress 0.1837223
#> Run 15 stress 0.18591
#> Run 16 stress 0.1874962
#> Run 17 stress 0.1823433
#> Run 18 stress 0.1848213
#> Run 19 stress 0.184988
#> Run 20 stress 0.1831431
#> *** Best solution was not repeated -- monoMDS stopping criteria:
#> 15: no. of iterations >= maxit
#> 5: stress ratio > sratmax
# \donttest{
library(patchwork)
physeq <- data_fungi_mini
res_tsne <- tsne_pq(data_fungi_mini)
df_umap_tsne <- df_umap
df_umap_tsne$x_tsne <- res_tsne$Y[, 1]
df_umap_tsne$y_tsne <- res_tsne$Y[, 2]
((ggplot(df_umap, aes(x = x_umap, y = y_umap, col = Height)) +
geom_point(size = 2) +
ggtitle("UMAP")) + (plot_ordination(physeq,
ordination =
ordinate(physeq, method = "PCoA", distance = "bray"), color = "Height"
)) +
ggtitle("PCoA")) /
((ggplot(df_umap_tsne, aes(x = x_tsne, y = y_tsne, col = Height)) +
geom_point(size = 2) +
ggtitle("tsne")) +
(plot_ordination(physeq,
ordination = ordinate(physeq, method = "NMDS", distance = "bray"),
color = "Height"
) +
ggtitle("NMDS"))) +
patchwork::plot_layout(guides = "collect")
#> Square root transformation
#> Wisconsin double standardization
#> Run 0 stress 0.1842444
#> Run 1 stress 0.1864672
#> Run 2 stress 0.1816642
#> ... New best solution
#> ... Procrustes: rmse 0.06035663 max resid 0.2763356
#> Run 3 stress 0.1805408
#> ... New best solution
#> ... Procrustes: rmse 0.0696832 max resid 0.2635835
#> Run 4 stress 0.1918613
#> Run 5 stress 0.1837193
#> Run 6 stress 0.1856894
#> Run 7 stress 0.1854912
#> Run 8 stress 0.1855901
#> Run 9 stress 0.1828179
#> Run 10 stress 0.1797561
#> ... New best solution
#> ... Procrustes: rmse 0.05144056 max resid 0.2994319
#> Run 11 stress 0.1861731
#> Run 12 stress 0.1861394
#> Run 13 stress 0.1881243
#> Run 14 stress 0.1837223
#> Run 15 stress 0.18591
#> Run 16 stress 0.1874962
#> Run 17 stress 0.1823433
#> Run 18 stress 0.1848213
#> Run 19 stress 0.184988
#> Run 20 stress 0.1831431
#> *** Best solution was not repeated -- monoMDS stopping criteria:
#> 15: no. of iterations >= maxit
#> 5: stress ratio > sratmax
 df_uwot <- umap_pq(data_fungi_mini, pkg = "uwot")
#> Taxa are now in columns.
#> Taxa are now in rows.
#> Joining with `by = join_by(Sample)`
#> Joining with `by = join_by(Sample)`
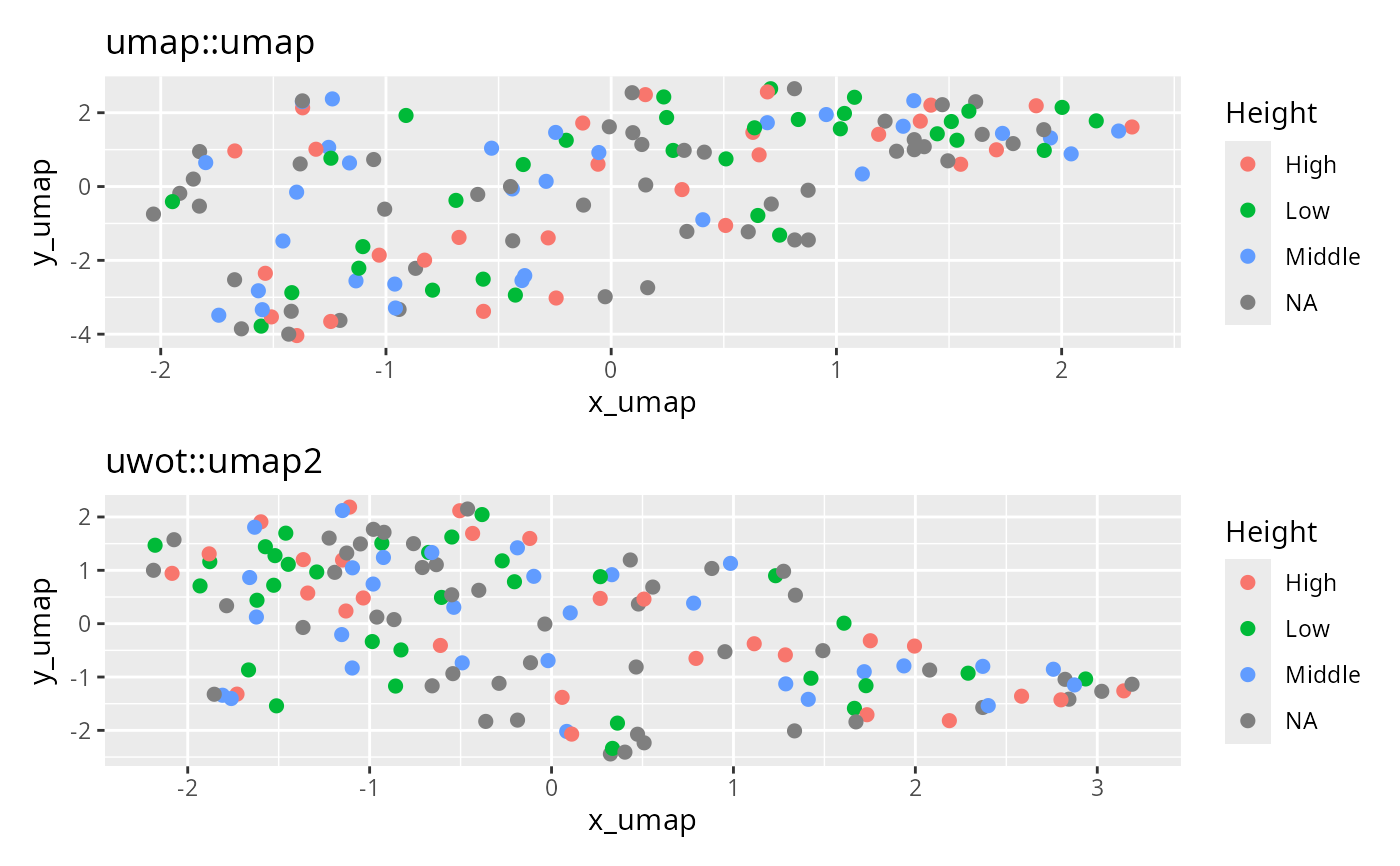
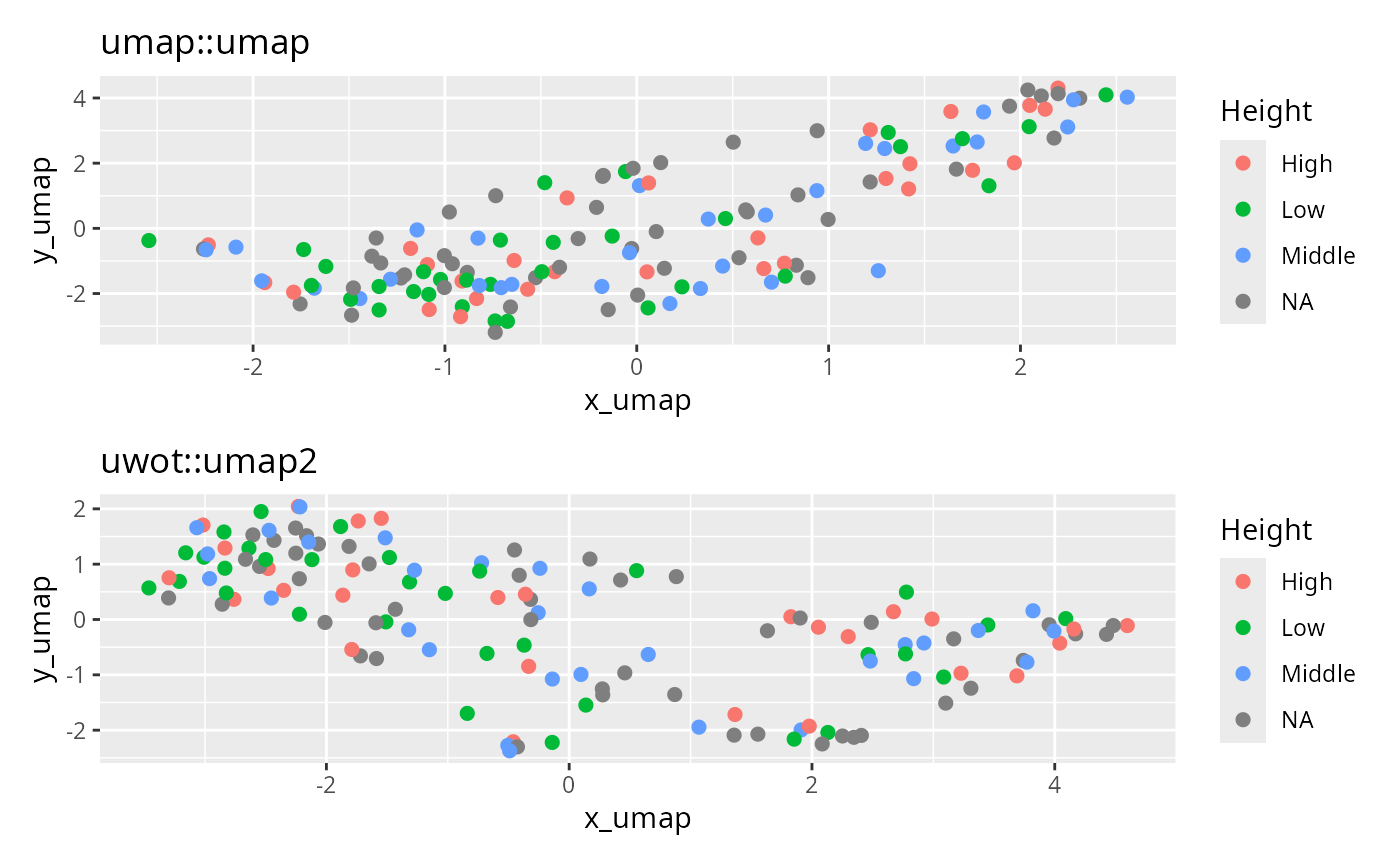
(ggplot(df_umap, aes(x = x_umap, y = y_umap, col = Height)) +
geom_point(size = 2) +
ggtitle("umap::umap")) /
(ggplot(df_uwot, aes(x = x_umap, y = y_umap, col = Height)) +
geom_point(size = 2) +
ggtitle("uwot::umap2"))
df_uwot <- umap_pq(data_fungi_mini, pkg = "uwot")
#> Taxa are now in columns.
#> Taxa are now in rows.
#> Joining with `by = join_by(Sample)`
#> Joining with `by = join_by(Sample)`
(ggplot(df_umap, aes(x = x_umap, y = y_umap, col = Height)) +
geom_point(size = 2) +
ggtitle("umap::umap")) /
(ggplot(df_uwot, aes(x = x_umap, y = y_umap, col = Height)) +
geom_point(size = 2) +
ggtitle("uwot::umap2"))
 # }
# }
