Box/Violin plots for between-subjects comparisons of Hill Number
Source:R/plot_functions.R
ggbetween_pq.RdNote that contrary to hill_pq(), this function does not take into
account for difference in the number of sequences per samples/modalities.
You may use rarefy_by_sample = TRUE if the mean number of sequences per
samples differs among modalities.
Basically a wrapper of function ggstatsplot::ggbetweenstats() for
object of class phyloseq
Usage
ggbetween_pq(
physeq,
fact,
one_plot = FALSE,
rarefy_by_sample = FALSE,
rngseed = FALSE,
verbose = TRUE,
...
)Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- fact
(required) The variable to test. Must be present in the
sam_dataslot of the physeq object.- one_plot
(logical, default FALSE) If TRUE, return a unique plot with the three plot inside using the patchwork package.
- rarefy_by_sample
(logical, default FALSE) If TRUE, rarefy samples using
phyloseq::rarefy_even_depth()function- rngseed
(Optional). A single integer value passed to
phyloseq::rarefy_even_depth(), which is used to fix a seed for reproducibly random number generation (in this case, reproducibly random subsampling). If set to FALSE, then no fiddling with the RNG seed is performed, and it is up to the user to appropriately call set.seed beforehand to achieve reproducible results. Default is FALSE.- verbose
(logical). If TRUE, print additional information.
- ...
Additional arguments passed on to
ggstatsplot::ggbetweenstats()function.
Value
Either an unique ggplot2 object (if one_plot is TRUE) or a list of 3 ggplot2 plot:
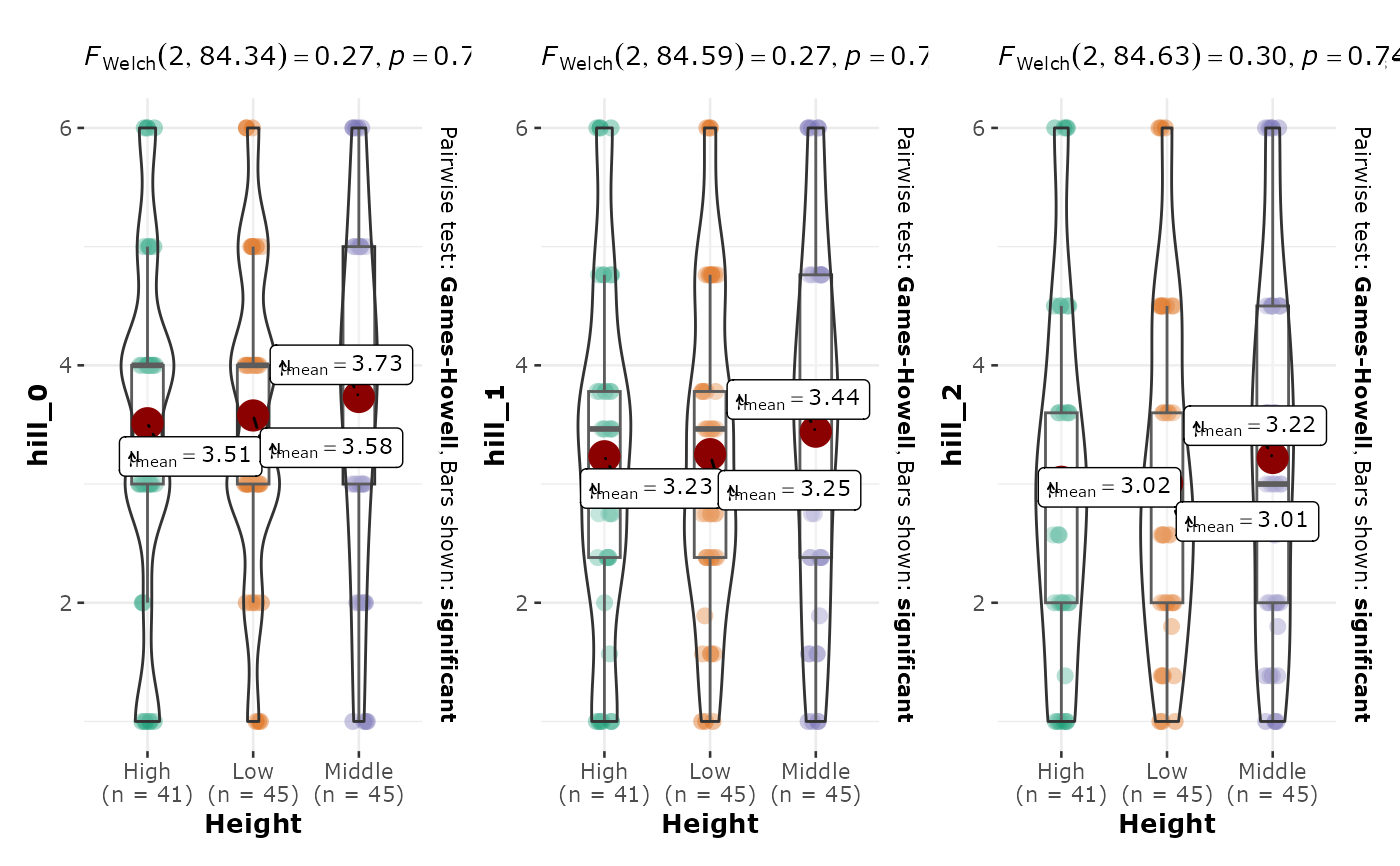
plot_Hill_0 : the ggbetweenstats of Hill number 0 (= species richness) against the variable fact
plot_Hill_1 : the ggbetweenstats of Hill number 1 (= Shannon index) against the variable fact
plot_Hill_2 : the ggbetweenstats of Hill number 2 (= Simpson index) against the variable fact
Details
This function is mainly a wrapper of the work of others.
Please make a reference to ggstatsplot::ggbetweenstats() if you
use this function.
Examples
# \donttest{
if (requireNamespace("ggstatsplot")) {
p <- ggbetween_pq(data_fungi, fact = "Time", p.adjust.method = "BH")
p[[1]]
ggbetween_pq(data_fungi, fact = "Height", one_plot = TRUE)
ggbetween_pq(data_fungi, fact = "Height", one_plot = TRUE, rarefy_by_sample = TRUE)
}
#> Warning: The mean number of sequences per samples vary across modalities of the variable 'Time' You should use rarefy_by_sample = TRUE or try hill_pq() with correction_for_sample_size = TRUE
#> You set `rngseed` to FALSE. Make sure you've set & recorded
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> ...
#> You set `rngseed` to FALSE. Make sure you've set & recorded
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> ...
#> 1020OTUs were removed because they are no longer
#> present in any sample after random subsampling
#> ...
#> All modality were undoubtedly rarefy in the physeq object.
 # }
# }
