Plot the partition the variation of a phyloseq object
Source:R/plot_functions.R
plot_var_part_pq.RdGraphical representation of the partition of variation obtain with var_par_pq().
Usage
plot_var_part_pq(
res_varpart,
cutoff = 0,
digits = 1,
digits_quantile = 2,
fill_bg = c("seagreen3", "mediumpurple", "blue", "orange"),
show_quantiles = FALSE,
filter_quantile_zero = TRUE,
show_dbrda_signif = FALSE,
show_dbrda_signif_pval = 0.05,
alpha = 63,
id.size = 1.2,
min_prop_pval_signif_dbrda = 0.95
)Arguments
- res_varpart
(required) the result of the functions
var_par_pq()orvar_par_rarperm_pq()- cutoff
The values below cutoff will not be displayed.
- digits
The number of significant digits.
- digits_quantile
The number of significant digits for quantile.
- fill_bg
Fill colours of ellipses.
- show_quantiles
Do quantiles are printed ?
- filter_quantile_zero
Do we filter out value with quantile encompassing the zero value?
- show_dbrda_signif
Do dbrda significance for each component is printed using *?
- show_dbrda_signif_pval
(float,
[0:1]) The value under which the dbrda is considered significant.- alpha
(int,
[0:255]) Transparency of the fill colour.- id.size
A numerical value giving the character expansion factor for the names of circles or ellipses.
- min_prop_pval_signif_dbrda
(float,
[0:1]) Only used if using the result ofvar_par_rarperm_pq()function. The * for dbrda_signif is only add if at leastmin_prop_pval_signif_dbrdaof permutations show significance.
Details
This function is mainly a wrapper of the work of others.
Please make a reference to vegan::varpart() if you
use this function.
Examples
# \donttest{
if (requireNamespace("vegan")) {
data_fungi_woNA <- subset_samples(data_fungi, !is.na(Time) & !is.na(Height))
res_var_9 <- var_par_rarperm_pq(
data_fungi_woNA,
list_component = list(
"Time" = c("Time"),
"Size" = c("Height", "Diameter")
),
nperm = 9,
dbrda_computation = TRUE
)
res_var_2 <- var_par_rarperm_pq(
data_fungi_woNA,
list_component = list(
"Time" = c("Time"),
"Size" = c("Height", "Diameter")
),
nperm = 2,
dbrda_computation = TRUE
)
res_var0 <- var_par_pq(data_fungi_woNA,
list_component = list(
"Time" = c("Time"),
"Size" = c("Height", "Diameter")
),
dbrda_computation = TRUE
)
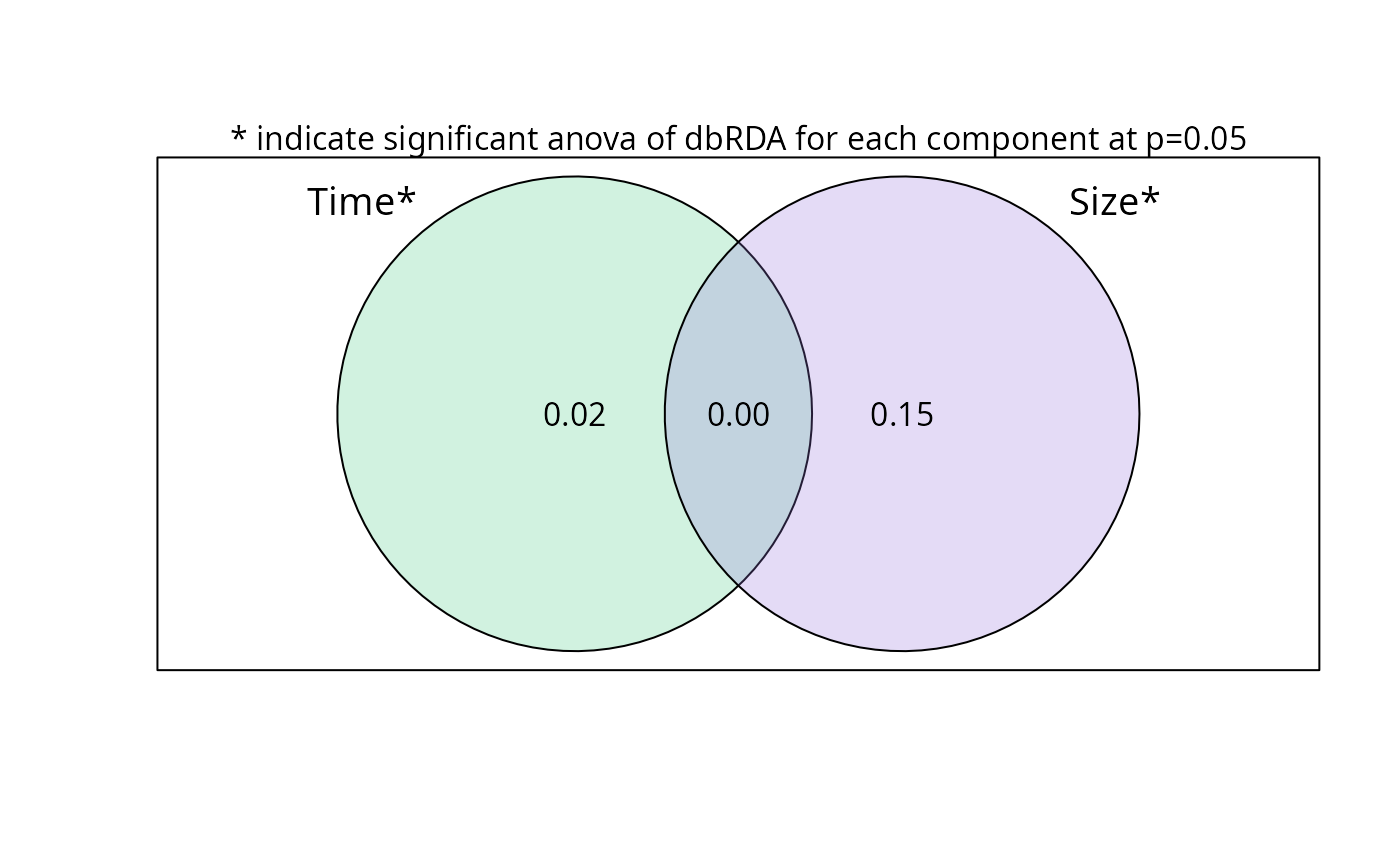
plot_var_part_pq(res_var0, digits_quantile = 2, show_dbrda_signif = TRUE)
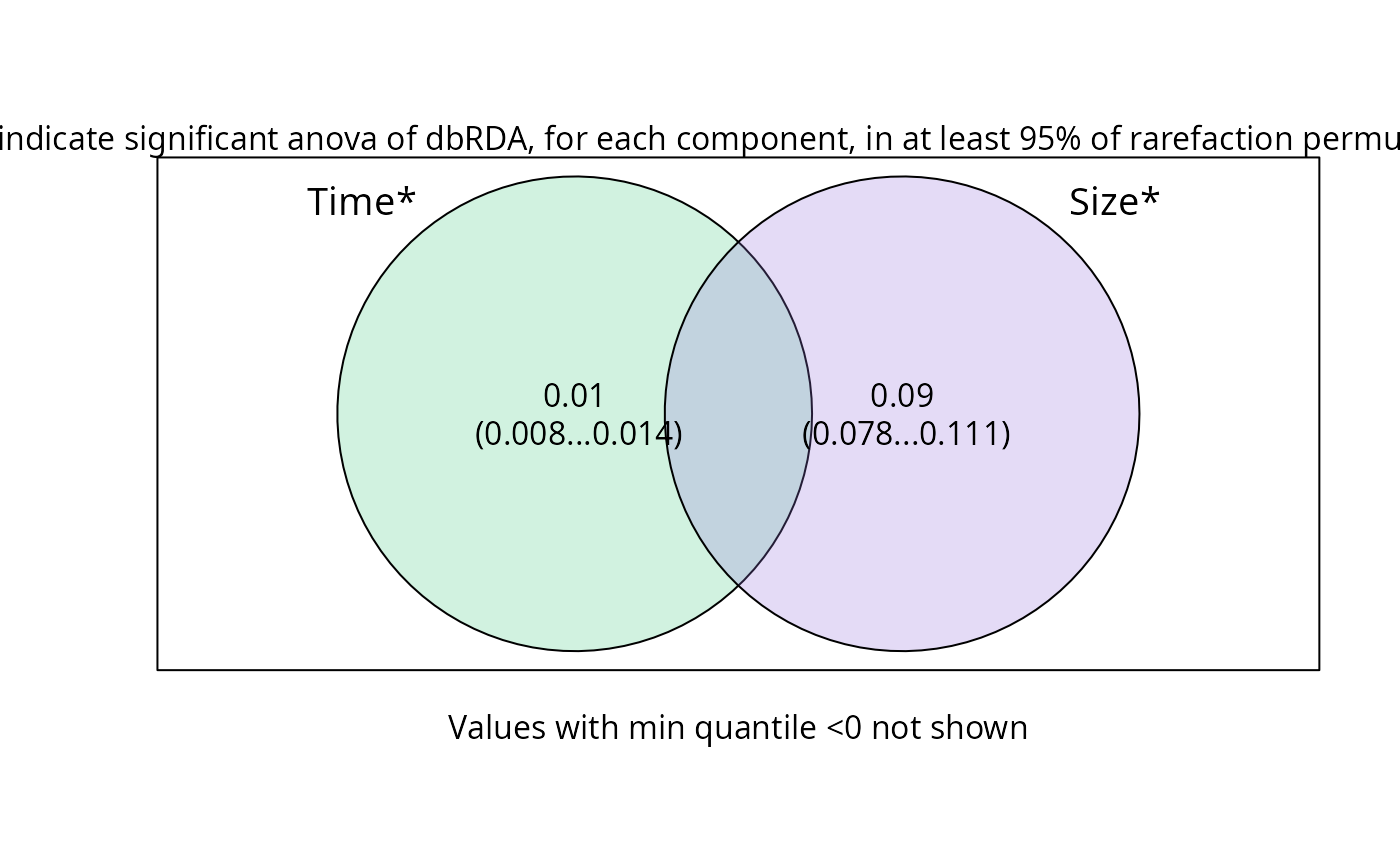
plot_var_part_pq(res_var_9,
digits_quantile = 2, show_quantiles = TRUE,
show_dbrda_signif = TRUE
)
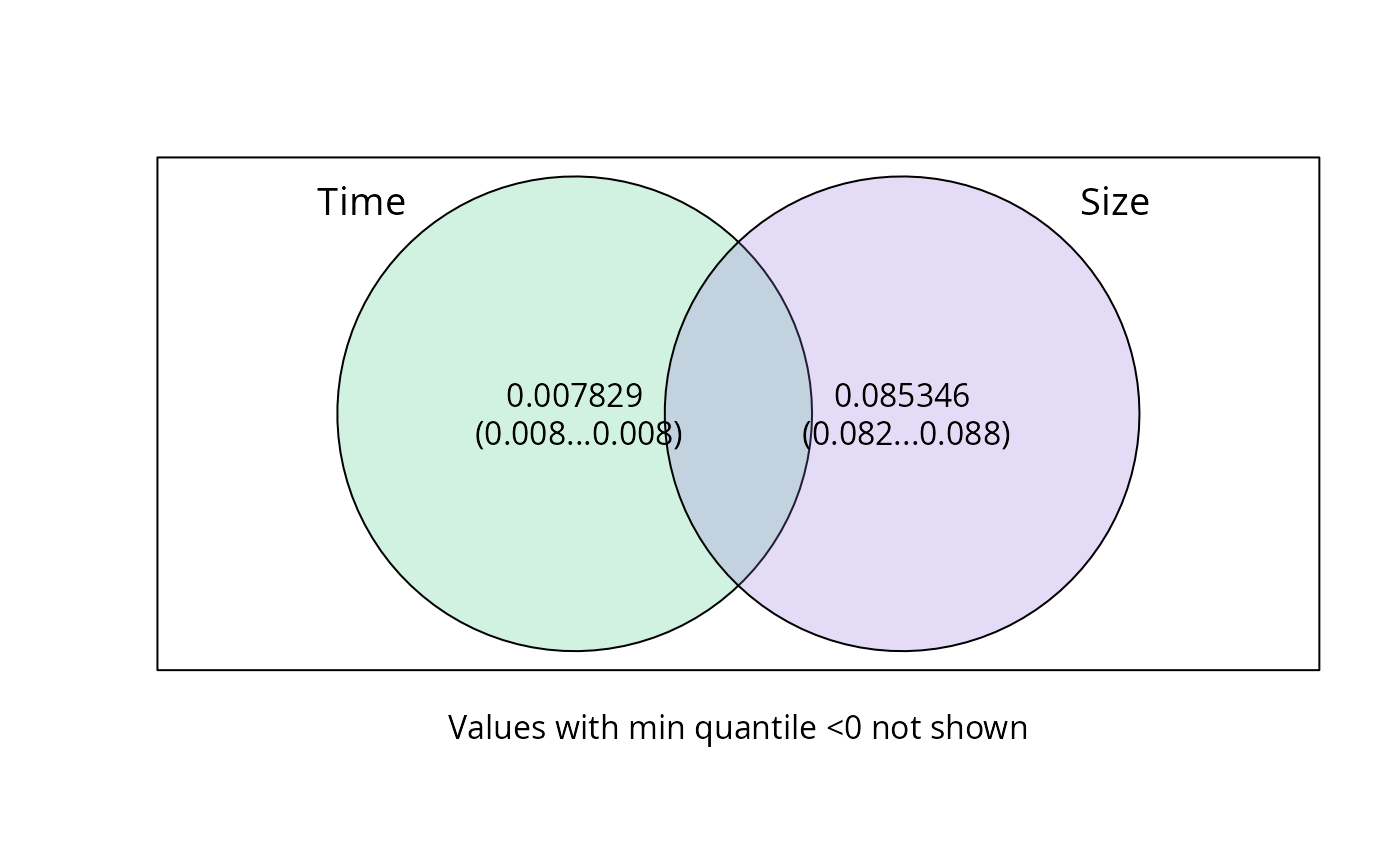
plot_var_part_pq(
res_var_2,
digits = 5,
digits_quantile = 2,
cutoff = 0,
show_quantiles = TRUE
)
}
#>
|
| | 0%
|
|====== | 11%
|
|=========== | 22%
|
|================= | 33%
|
|====================== | 44%
|
|============================ | 56%
|
|================================= | 67%
|
|======================================= | 78%
|
|============================================ | 89%
|
|==================================================| 100%
|
| | 0%
|
|========================= | 50%
|
|==================================================| 100%
 # }
# }
