Partition the Variation of a phyloseq object with rarefaction permutations
Source:R/beta_div_test.R
var_par_rarperm_pq.RdThis is an extension of the function var_par_pq(). The main addition is
the computation of nperm permutations with rarefaction even depth by
sample. The return object
Usage
var_par_rarperm_pq(
physeq,
list_component,
dist_method = "bray",
nperm = 99,
quantile_prob = 0.975,
dbrda_computation = FALSE,
dbrda_signif_pval = 0.05,
sample.size = min(sample_sums(physeq)),
verbose = FALSE,
progress_bar = TRUE
)Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- list_component
(required) A named list of 2, 3 or four vectors with names from the
@sam_dataslot.- dist_method
(default "bray") the distance used. See
phyloseq::distance()for all available distances or runphyloseq::distanceMethodList(). For aitchison and robust.aitchison distance,vegan::vegdist()function is directly used.#' @param fill_bg- nperm
(int) The number of permutations to perform.
- quantile_prob
(float,
[0:1]) the value to compute the quantile. Minimum quantile is compute using 1-quantile_prob.- dbrda_computation
(logical) Do dbrda computations are runned for each individual component (each name of the list component) ?
- dbrda_signif_pval
(float,
[0:1]) The value under which the dbrda is considered significant.- sample.size
(int) A single integer value equal to the number of reads being simulated, also known as the depth. See
phyloseq::rarefy_even_depth().- verbose
(logical). If TRUE, print additional information.
- progress_bar
(logical, default TRUE) Do we print progress during the calculation?
Value
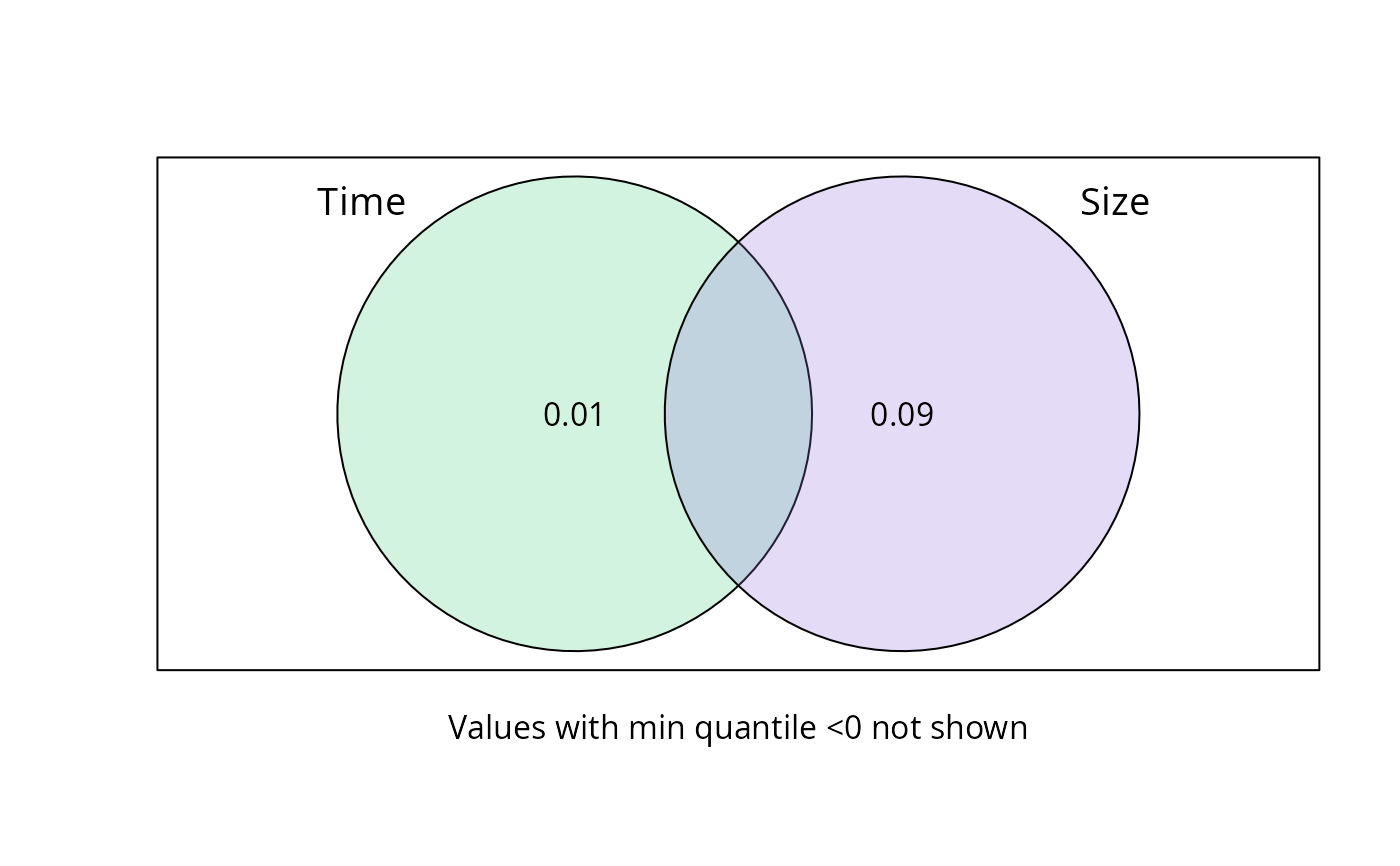
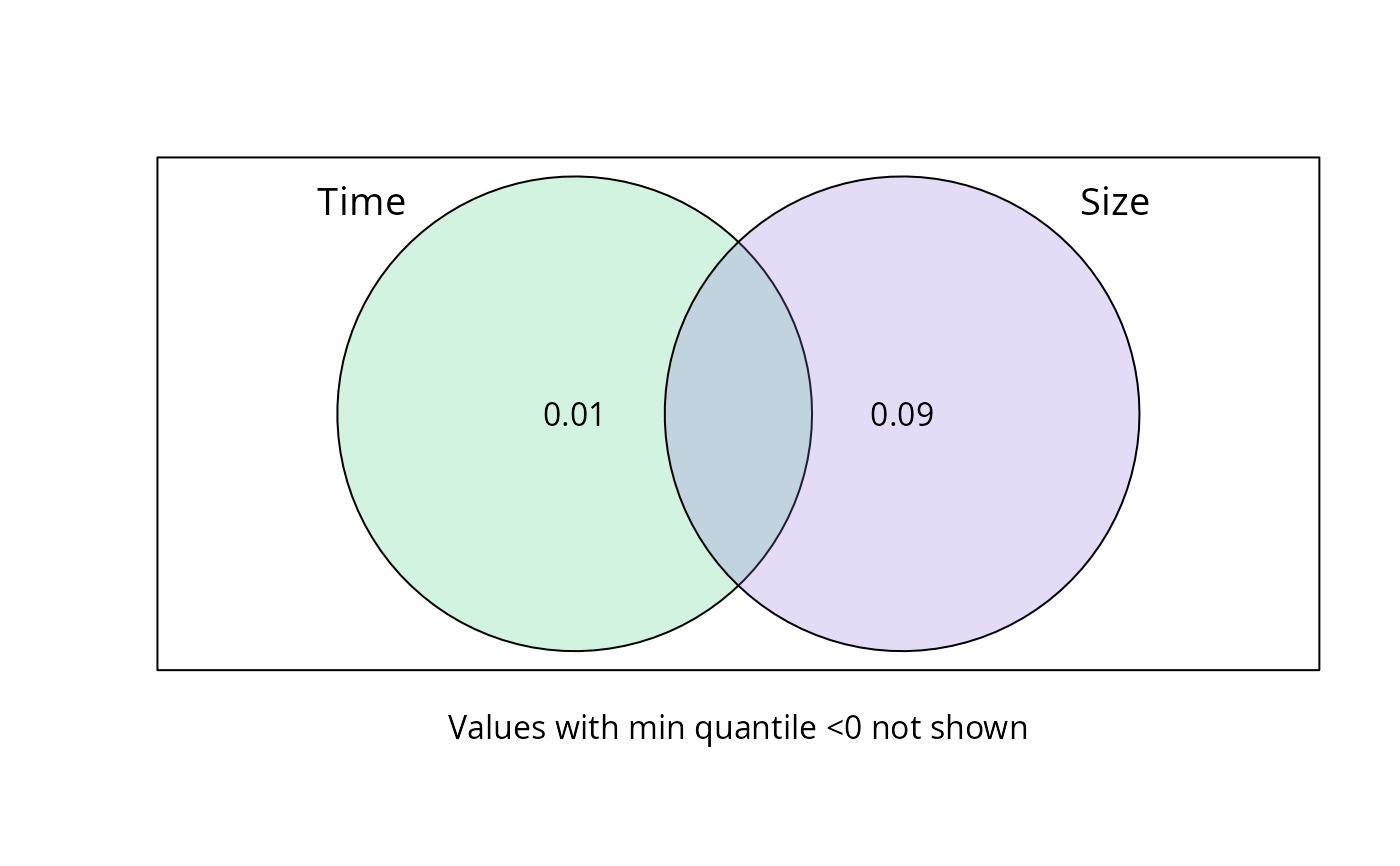
A list of class varpart with additional information in the
$part$indfract part. Adj.R.square is the mean across permutation.
Adj.R.squared_quantil_min and Adj.R.squared_quantil_max represent
the quantile values of adjusted R squared
Details
This function is mainly a wrapper of the work of others.
Please make a reference to vegan::varpart() if you
use this function.
Examples
# \donttest{
if (requireNamespace("vegan")) {
data_fungi_woNA <- subset_samples(data_fungi, !is.na(Time) & !is.na(Height))
res_var_9 <- var_par_rarperm_pq(
data_fungi_woNA,
list_component = list(
"Time" = c("Time"),
"Size" = c("Height", "Diameter")
),
nperm = 9,
dbrda_computation = TRUE
)
plot_var_part_pq(res_var_9)
res_var_2 <- var_par_rarperm_pq(
data_fungi_woNA,
list_component = list(
"Time" = c("Time"),
"Size" = c("Height", "Diameter")
),
nperm = 2,
dbrda_computation = TRUE
)
plot_var_part_pq(res_var_2)
}
#>
|
| | 0%
|
|====== | 11%
|
|=========== | 22%
|
|================= | 33%
|
|====================== | 44%
|
|============================ | 56%
|
|================================= | 67%
|
|======================================= | 78%
|
|============================================ | 89%
|
|==================================================| 100%
 #>
|
| | 0%
|
|========================= | 50%
|
|==================================================| 100%
#>
|
| | 0%
|
|========================= | 50%
|
|==================================================| 100%
 # }
# }
