Hill numbers are the number of equiprobable species giving the same diversity value as the observed distribution.
Note that contrary to hill_pq(), this function does not take into
account for difference in the number of sequences per samples/modalities.
You may use rarefy_by_sample = TRUE if the mean number of sequences per
samples differs among modalities.
Usage
psmelt_samples_pq(
physeq,
hill_scales = c(0, 1, 2),
filter_zero = TRUE,
rarefy_by_sample = FALSE,
rngseed = FALSE,
verbose = TRUE,
taxa_ranks = NULL
)Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- hill_scales
(a vector of integer) The list of q values to compute the hill number H^q. If Null, no hill number are computed. Default value compute the Hill number 0 (Species richness), the Hill number 1 (exponential of Shannon Index) and the Hill number 2 (inverse of Simpson Index).
- filter_zero
(logical, default TRUE) Do we filter non present OTU from samples ? For the moment, this has no effect on the result because the dataframe is grouped by samples with abundance summed across OTU.
- rarefy_by_sample
(logical, default FALSE) If TRUE, rarefy samples using
phyloseq::rarefy_even_depth()function.- rngseed
(Optional). A single integer value passed to
phyloseq::rarefy_even_depth(), which is used to fix a seed for reproducibly random number generation (in this case, reproducibly random subsampling). If set to FALSE, then no fiddling with the RNG seed is performed, and it is up to the user to appropriately call set.seed beforehand to achieve reproducible results. Default is FALSE.- verbose
(logical). If TRUE, print additional information.
- taxa_ranks
A vector of taxonomic ranks. For examples c("Family","Genus"). If taxa ranks is not set (default value = NULL), taxonomic information are not present in the resulting tibble.
Value
A tibble with a row for each sample. Columns provide information
from sam_data slot as well as hill numbers, Abundance (nb of sequences),
and Abundance_log10 (log10(1+Abundance)).
Examples
if (requireNamespace("ggstatsplot")) {
psm_tib <- psmelt_samples_pq(data_fungi_mini, hill_scales = c(0, 2, 7))
ggstatsplot::ggbetweenstats(psm_tib, Height, Hill_0)
ggstatsplot::ggbetweenstats(psm_tib, Height, Hill_7)
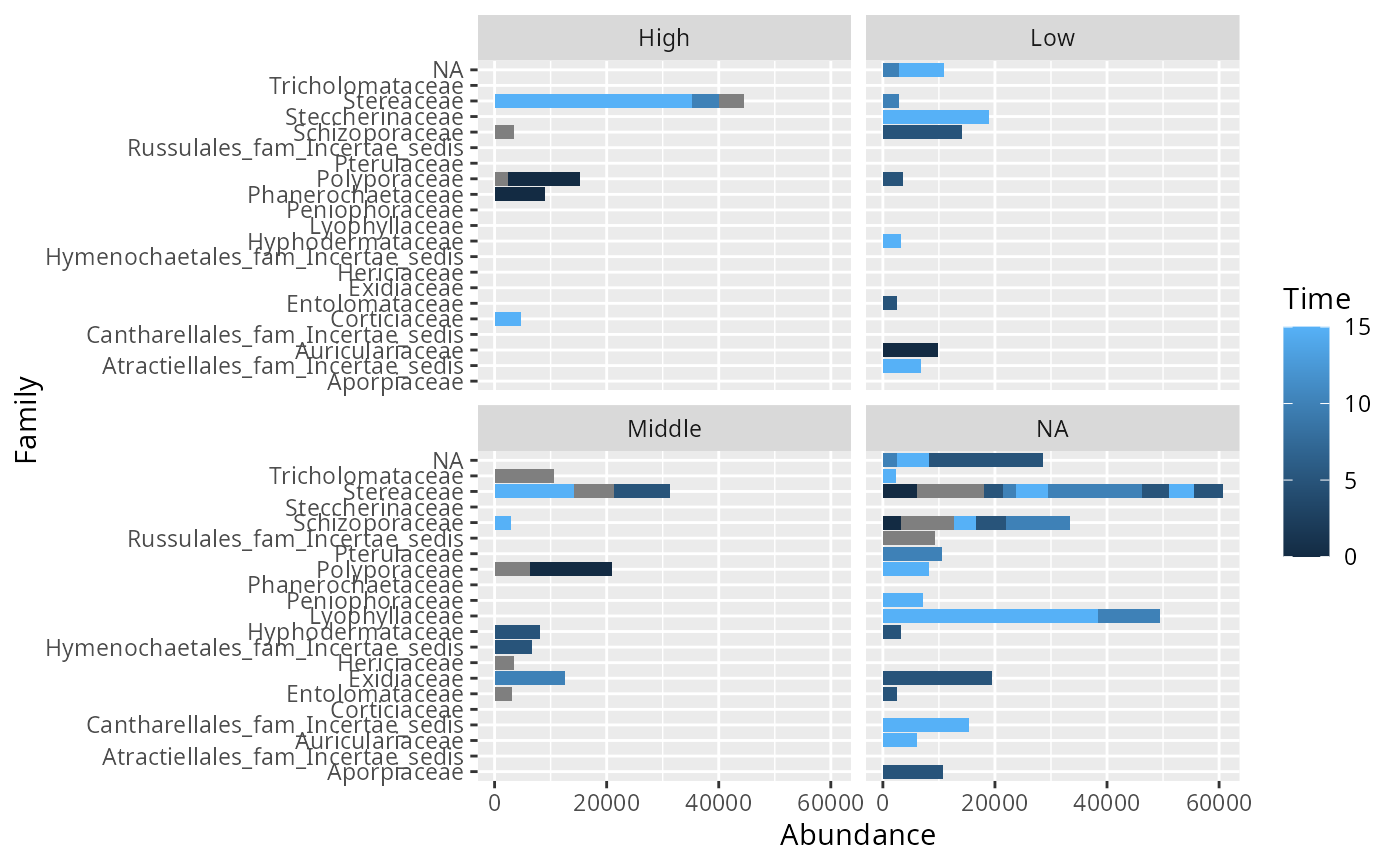
psm_tib_tax <- psmelt_samples_pq(data_fungi_mini, taxa_ranks = c("Class", "Family"))
ggplot(filter(psm_tib_tax, Abundance > 2000), aes(y = Family, x = Abundance, fill = Time)) +
geom_bar(stat = "identity") +
facet_wrap(~Height)
}
#> Joining with `by = join_by(Sample)`
#> Joining with `by = join_by(Sample)`