Rarefy (equalize) the number of samples per modality of a factor
Source:R/dada_phyloseq.R
rarefy_sample_count_by_modality.RdThis function randomly draw the same number of samples for each modality of factor.
It is usefull to dissentangle the effect of different number of samples per modality
on diversity. Internally used in accu_plot_balanced_modality().
Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- fact
(required) The variable to rarefy. Must be present in the
sam_dataslot of the physeq object.- rngseed
(Optional). A single integer value passed to set.seed, which is used to fix a seed for reproducibly random number generation (in this case, reproducibly random subsampling). If set to FALSE, then no iddling with the RNG seed is performed, and it is up to the user to appropriately call
- verbose
(logical). If TRUE, print additional information.
Value
A new phyloseq-class object.
Examples
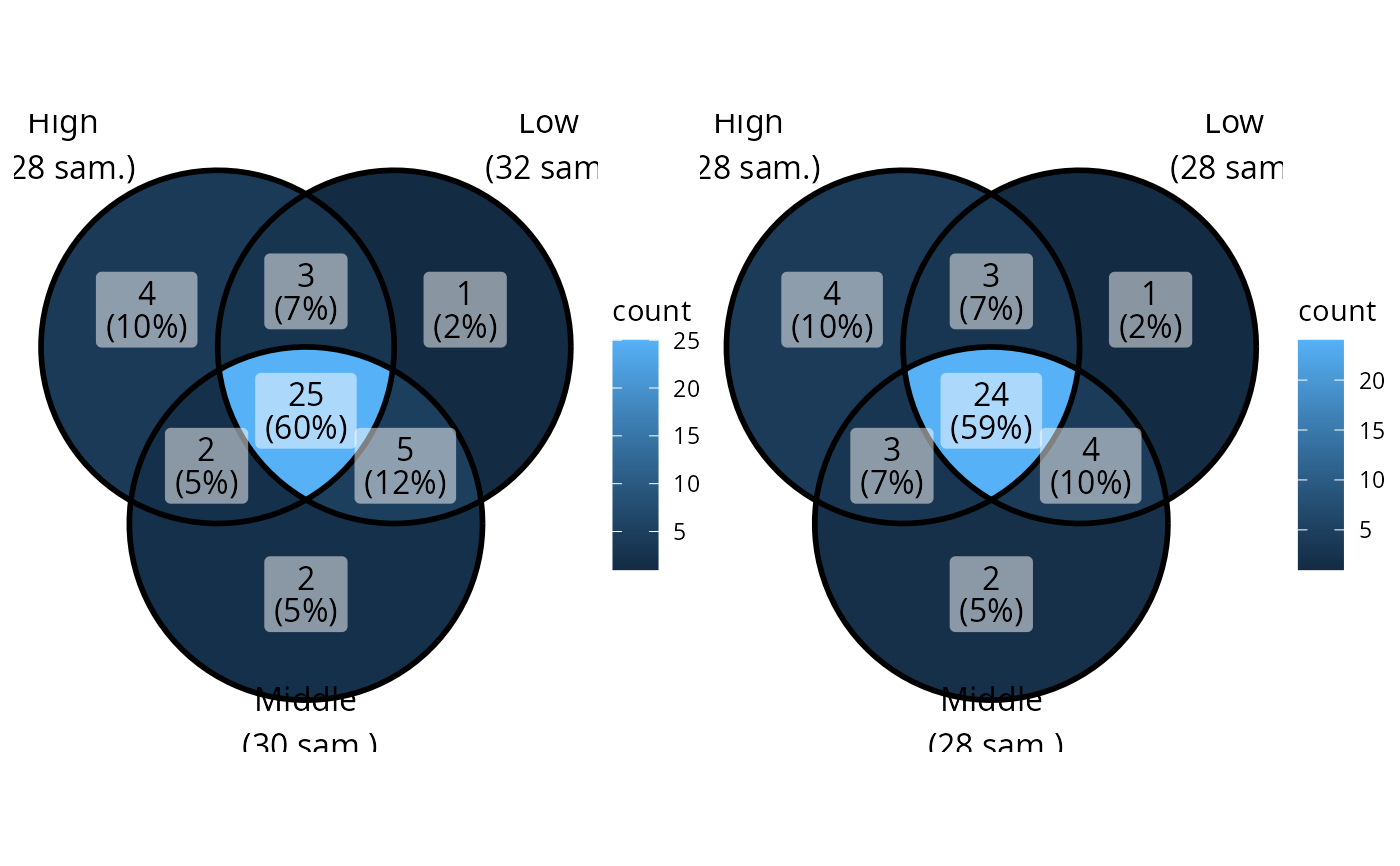
table(data_fungi_mini@sam_data$Height)
#>
#> High Low Middle
#> 28 32 30
data_fungi_mini2 <- rarefy_sample_count_by_modality(data_fungi_mini, "Height")
#> You set `rngseed` to FALSE. Make sure you've set & recorded
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> ...
table(data_fungi_mini2@sam_data$Height)
#>
#> High Low Middle
#> 28 28 28
if (requireNamespace("patchwork")) {
ggvenn_pq(data_fungi_mini, "Height") + ggvenn_pq(data_fungi_mini2, "Height")
}
#> 47 were discarded due to NA in variable fact
#> Taxa are now in columns.
#> Cleaning suppress 11 taxa and 0 samples.
#> Cleaning suppress 11 taxa and 0 samples.
#> Cleaning suppress 11 taxa and 0 samples.
#> Taxa are now in columns.
#> Cleaning suppress 11 taxa and 0 samples.
#> Cleaning suppress 12 taxa and 0 samples.
#> Cleaning suppress 12 taxa and 0 samples.