Plot the distribution of sequences or ASV in one taxonomic levels
Source:R/plot_functions.R
tax_bar_pq.RdArguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- fact
Name of the factor to cluster samples by modalities. Need to be in
physeq@sam_data.- taxa
(default: 'Order') Name of the taxonomic rank of interest
- percent_bar
(default FALSE) If TRUE, the stacked bar fill all the space between 0 and 1. It just set position = "fill" in the
ggplot2::geom_bar()function- nb_seq
(logical; default TRUE) If set to FALSE, only the number of ASV is count. Concretely, physeq otu_table is transformed in a binary otu_table (each value different from zero is set to one)
Value
A ggplot2 plot with bar representing the number of sequence en each
taxonomic groups
Examples
data_fungi_ab <- subset_taxa_pq(data_fungi, taxa_sums(data_fungi) > 10000)
#> Cleaning suppress 0 taxa ( ) and 15 sample(s) ( BE9-006-B_S27_MERGED.fastq.gz / C21-NV1-M_S64_MERGED.fastq.gz / DJ2-008-B_S87_MERGED.fastq.gz / DY5-004-H_S97_MERGED.fastq.gz / DY5-004-M_S98_MERGED.fastq.gz / E9-009-B_S100_MERGED.fastq.gz / E9-009-H_S101_MERGED.fastq.gz / N22-001-B_S129_MERGED.fastq.gz / O20-X-B_S139_MERGED.fastq.gz / O21-007-M_S144_MERGED.fastq.gz / R28-008-H_S159_MERGED.fastq.gz / R28-008-M_S160_MERGED.fastq.gz / W26-001-M_S167_MERGED.fastq.gz / Y29-007-H_S182_MERGED.fastq.gz / Y29-007-M_S183_MERGED.fastq.gz ).
#> Number of non-matching ASV 0
#> Number of matching ASV 1420
#> Number of filtered-out ASV 1385
#> Number of kept ASV 35
#> Number of kept samples 170
tax_bar_pq(data_fungi_ab) + theme(legend.position = "none")
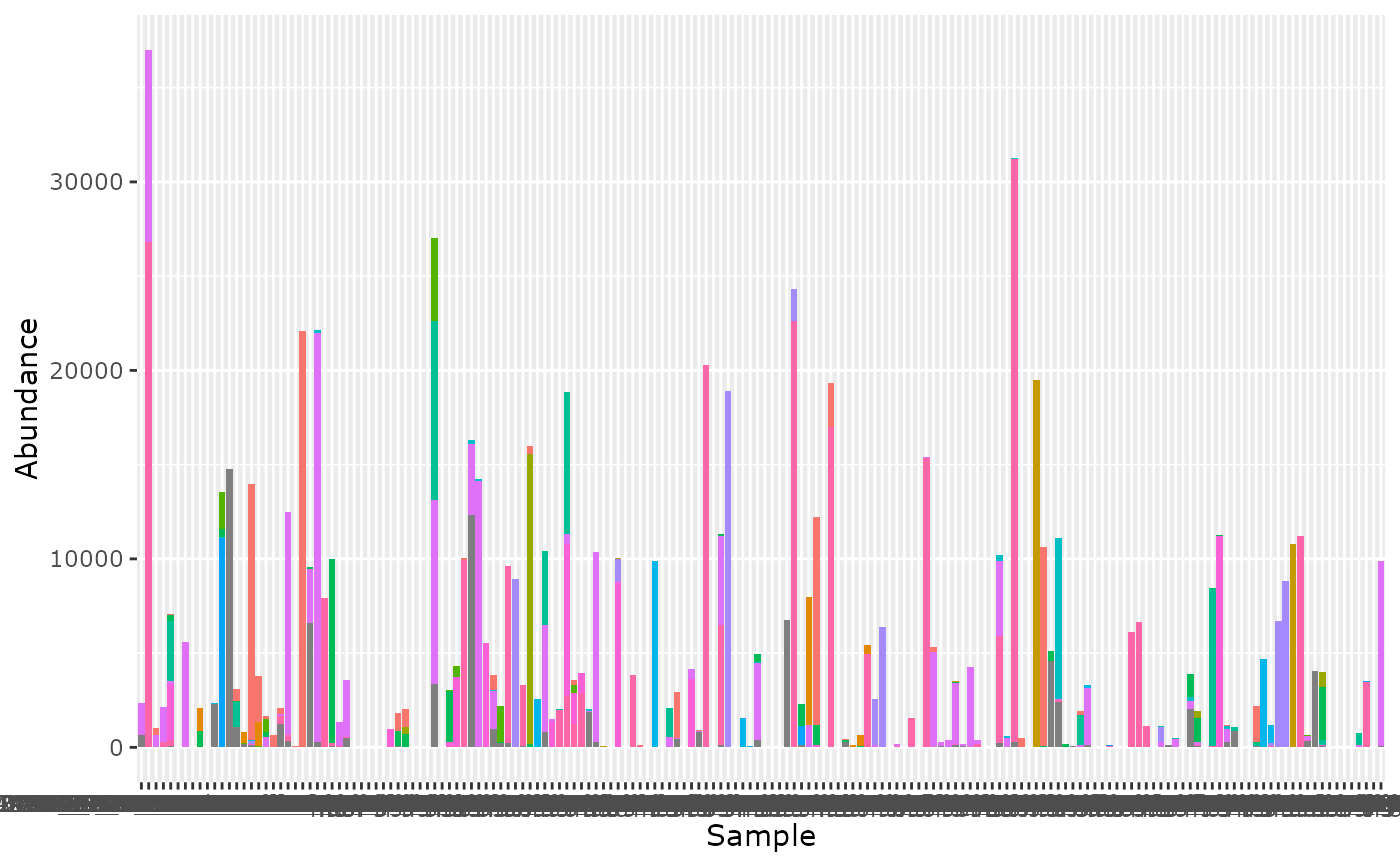 # \donttest{
tax_bar_pq(data_fungi_ab, taxa = "Class")
# \donttest{
tax_bar_pq(data_fungi_ab, taxa = "Class")
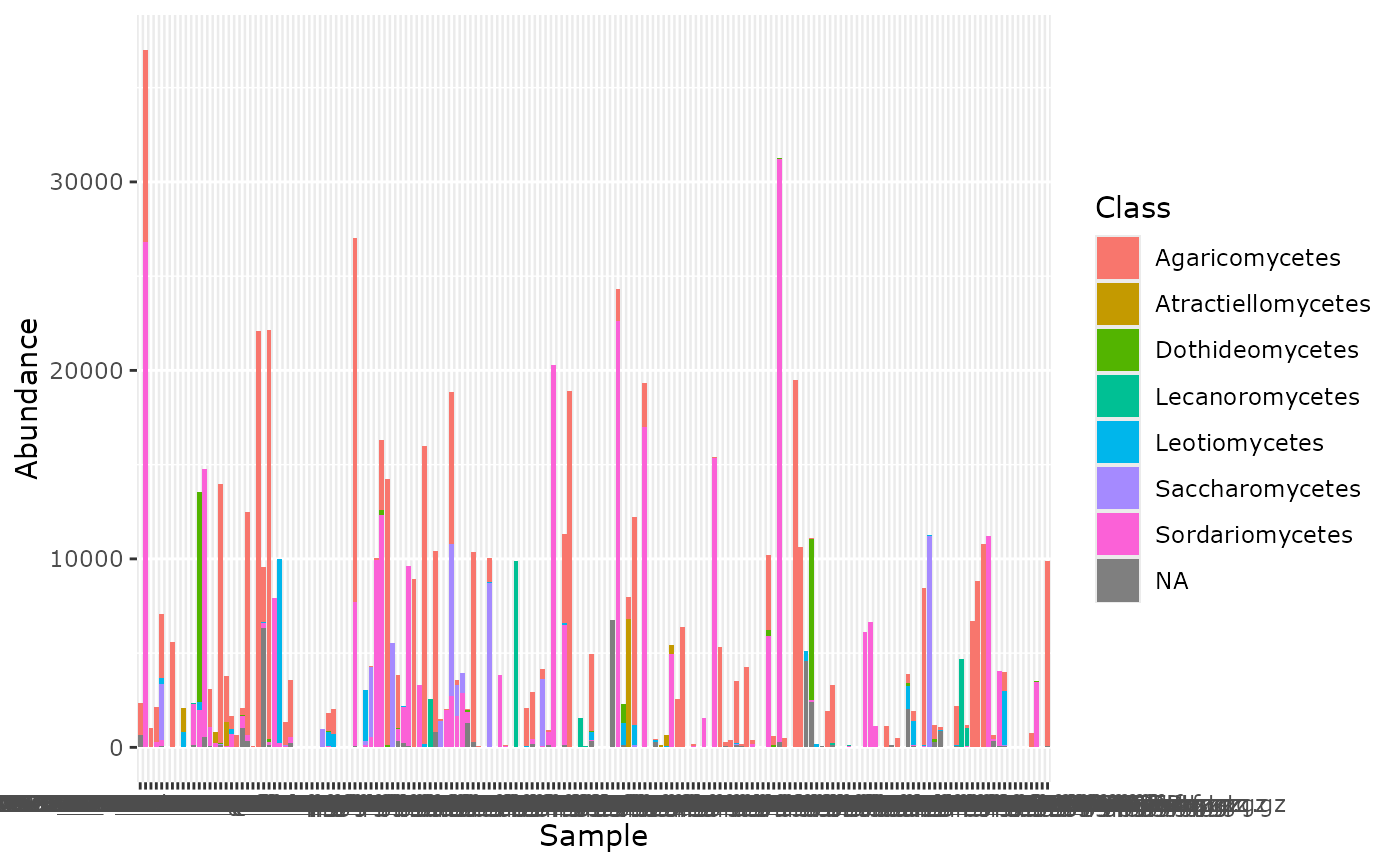 tax_bar_pq(data_fungi_ab, taxa = "Class", percent_bar = TRUE)
tax_bar_pq(data_fungi_ab, taxa = "Class", percent_bar = TRUE)
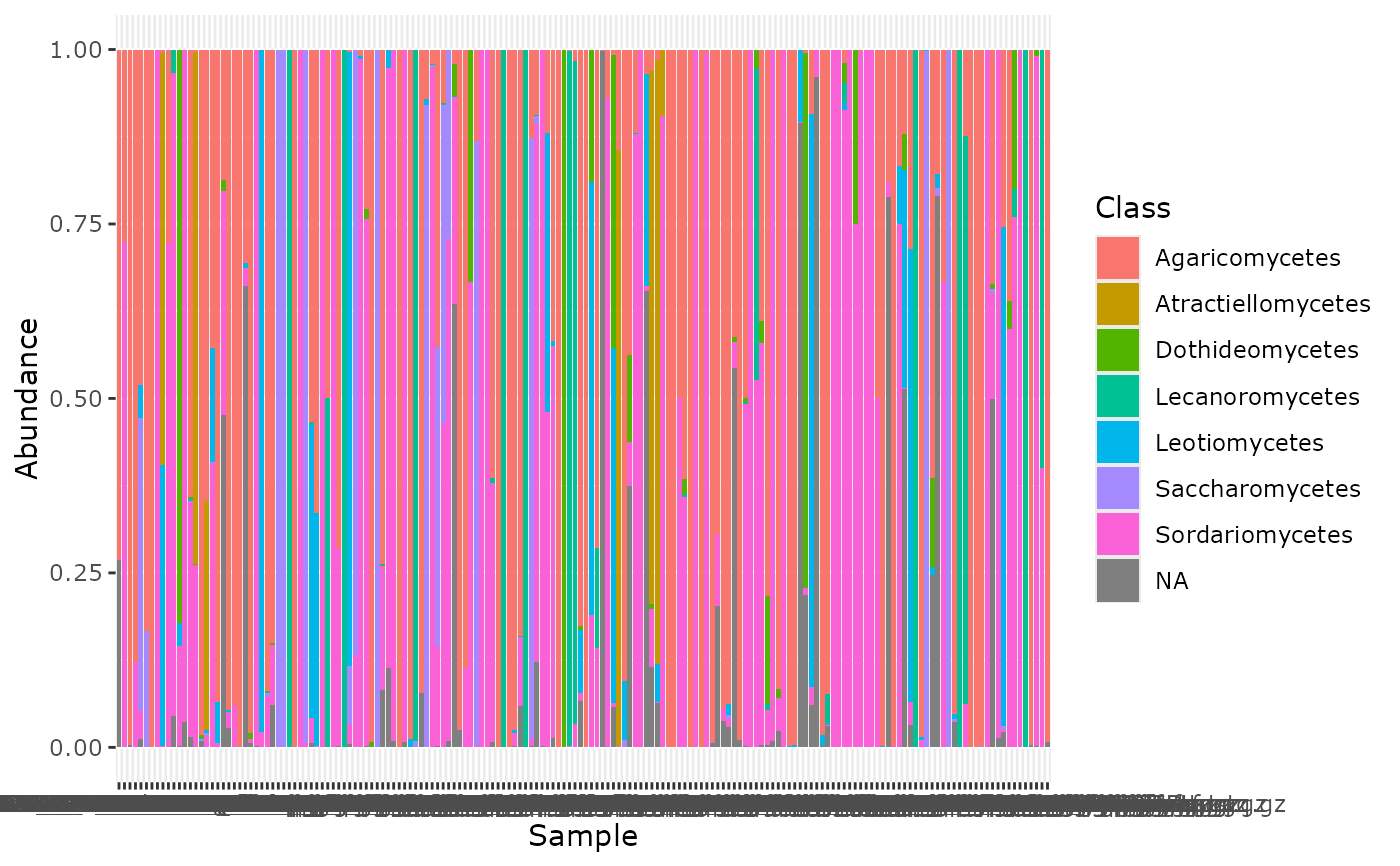 tax_bar_pq(data_fungi_ab, taxa = "Class", fact = "Time")
#> Warning: Removed 770 rows containing missing values or values outside the scale range
#> (`geom_bar()`).
tax_bar_pq(data_fungi_ab, taxa = "Class", fact = "Time")
#> Warning: Removed 770 rows containing missing values or values outside the scale range
#> (`geom_bar()`).
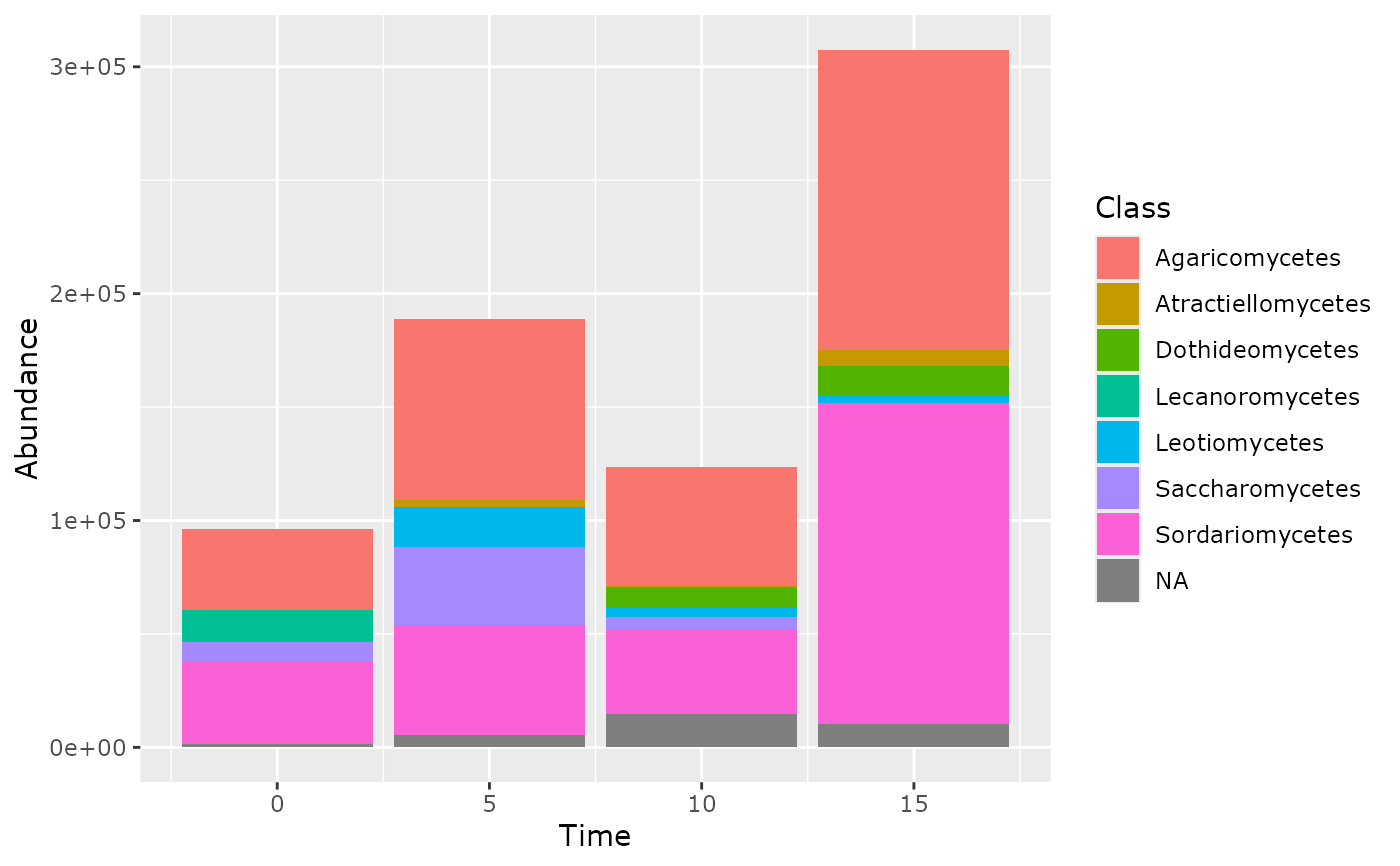 # }
# }
