Recluster sequences of an object of class physeq or cluster a list of DNA sequences using vsearch software
Source: R/vsearch.R
vsearch_clustering.RdUsage
vsearch_clustering(
physeq = NULL,
dna_seq = NULL,
nproc = 1,
id = 0.97,
vsearchpath = "vsearch",
tax_adjust = 0,
rank_propagation = FALSE,
vsearch_cluster_method = "--cluster_size",
vsearch_args = "--strand both",
keep_temporary_files = FALSE
)Arguments
- physeq
(required) a
phyloseq-classobject obtained using thephyloseqpackage.- dna_seq
You may directly use a character vector of DNA sequences in place of physeq args. When physeq is set, dna sequences take the value of
physeq@refseq- nproc
(default: 1) Set to number of cpus/processors to use for the clustering
- id
(default: 0.97) level of identity to cluster
- vsearchpath
(default: "vsearch") path to vsearch
- tax_adjust
(Default 0) See the man page of
merge_taxa_vec()for more details. To conserved the taxonomic rank of the most abundant ASV, set tax_adjust to 0 (default). For the moment only tax_adjust = 0 is robust- rank_propagation
(logical, default FALSE). Do we propagate the NA value from lower taxonomic rank to upper rank? See the man page of
merge_taxa_vec()for more details.- vsearch_cluster_method
(default: "–cluster_size) See other possible methods in the vsearch manual (e.g.
--cluster_sizeor--cluster_fast)--cluster_fast: Clusterize the fasta sequences in filename, automatically sort by decreasing sequence length beforehand.--cluster_size: Clusterize the fasta sequences in filename, automatically sort by decreasing sequence abundance beforehand.
- vsearch_args
(default : "–strand both") a one length character element defining other parameters to passed on to vsearch.
- keep_temporary_files
(logical, default: FALSE) Do we keep temporary files ?
temp.fasta (refseq in fasta or dna_seq sequences)
cluster.fasta (centroid if method = "vsearch")
temp.uc (clusters if method = "vsearch")
Details
This function use the merge_taxa_vec() function to
merge taxa into clusters. By default tax_adjust = 0. See the man page
of merge_taxa_vec().
This function is mainly a wrapper of the work of others. Please cite vsearch.
References
VSEARCH can be downloaded from https://github.com/torognes/vsearch. More information in the associated publication https://pubmed.ncbi.nlm.nih.gov/27781170.
Examples
# \donttest{
summary_plot_pq(data_fungi)
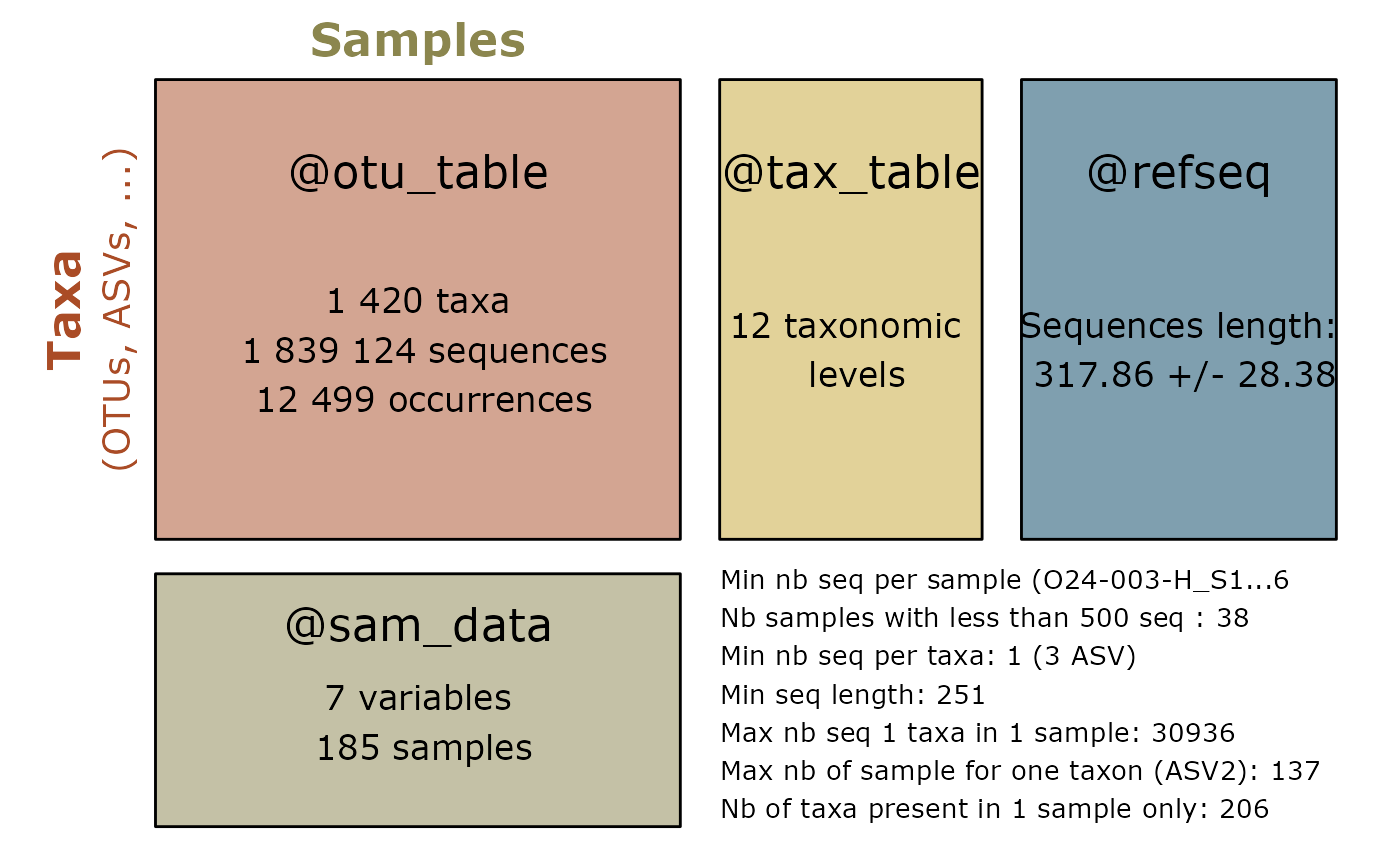 d_vs <- vsearch_clustering(data_fungi)
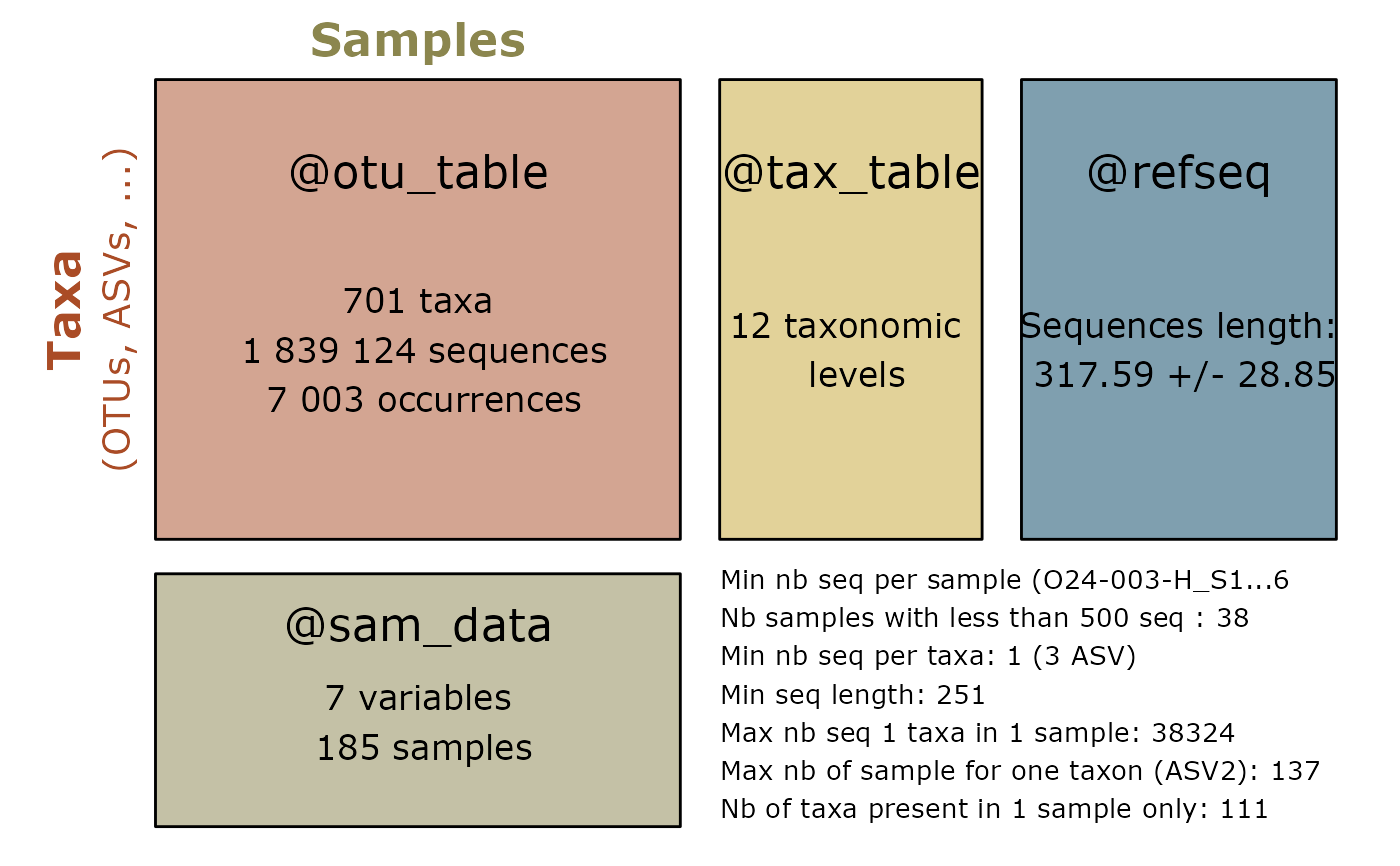
summary_plot_pq(d_vs)
d_vs <- vsearch_clustering(data_fungi)
summary_plot_pq(d_vs)
 # }
# }
